1BQR
 
 | | REDUCED PSEUDOAZURIN | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Inoue, T, Nishio, N, Hamanaka, S, Shimomura, T, Harada, S, Suzuki, S, Kohzuma, T, Shidara, S, Iwasaki, H, Kai, Y. | | Deposit date: | 1998-08-17 | | Release date: | 1999-08-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure determinations of oxidized and reduced pseudoazurins from Achromobacter cycloclastes. Concerted movement of copper site in redox forms with the rearrangement of hydrogen bond at a remote histidine.
J.Biol.Chem., 274, 1999
|
|
1GZB
 
 | | PEROXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PEROXIDASE, ... | | Authors: | Fukuyama, K, Kunishima, N, Amada, F. | | Deposit date: | 1996-11-13 | | Release date: | 1997-03-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pentacoordination of the heme iron of Arthromyces ramosus peroxidase shown by a 1.8 A resolution crystallographic study at pH 4.5.
FEBS Lett., 378, 1996
|
|
1CD9
 
 | | 2:2 COMPLEX OF G-CSF WITH ITS RECEPTOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (G-CSF RECEPTOR), PROTEIN (GRANULOCYTE COLONY-STIMULATING FACTOR) | | Authors: | Aritomi, M, Kunishima, N, Okamoto, T, Kuroki, R, Ota, Y, Morikawa, K. | | Deposit date: | 1999-03-08 | | Release date: | 2000-03-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Atomic structure of the GCSF-receptor complex showing a new cytokine-receptor recognition scheme.
Nature, 401, 1999
|
|
1ARW
 
 | | CRYSTAL STRUCTURES OF CYANIDE-AND TRIIODIDE-BOUND FORMS OF ARTHROMYCES RAMOSUS PEROXIDASE AT DIFFERENT PH VALUES. PERTURBATIONS OF ACTIVE SITE RESIDUES AND THEIR IMPLICATION IN ENZYME CATALYSIS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CYANIDE ION, ... | | Authors: | Fukuyama, K, Kunishima, N, Amada, F. | | Deposit date: | 1995-04-25 | | Release date: | 1996-01-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of cyanide- and triiodide-bound forms of Arthromyces ramosus peroxidase at different pH values. Perturbations of active site residues and their implication in enzyme catalysis.
J.Biol.Chem., 270, 1995
|
|
2ZKT
 
 | |
1ARV
 
 | | CRYSTAL STRUCTURES OF CYANIDE-AND TRIIODIDE-BOUND FORMS OF ARTHROMYCES RAMOSUS PEROXIDASE AT DIFFERENT PH VALUES. PERTURBATIONS OF ACTIVE SITE RESIDUES AND THEIR IMPLICATION IN ENZYME CATALYSIS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CYANIDE ION, ... | | Authors: | Fukuyama, K, Kunishima, N, Amada, F. | | Deposit date: | 1995-04-25 | | Release date: | 1996-01-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of cyanide- and triiodide-bound forms of Arthromyces ramosus peroxidase at different pH values. Perturbations of active site residues and their implication in enzyme catalysis.
J.Biol.Chem., 270, 1995
|
|
1ARU
 
 | | CRYSTAL STRUCTURES OF CYANIDE-AND TRIIODIDE-BOUND FORMS OF ARTHROMYCES RAMOSUS PEROXIDASE AT DIFFERENT PH VALUES. PERTURBATIONS OF ACTIVE SITE RESIDUES AND THEIR IMPLICATION IN ENZYME CATALYSIS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CYANIDE ION, ... | | Authors: | Fukuyama, K, Kunishima, N, Amada, F. | | Deposit date: | 1995-04-25 | | Release date: | 1996-01-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of cyanide- and triiodide-bound forms of Arthromyces ramosus peroxidase at different pH values. Perturbations of active site residues and their implication in enzyme catalysis.
J.Biol.Chem., 270, 1995
|
|
1J3W
 
 | |
1BQK
 
 | | OXIDIZED PSEUDOAZURIN | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Inoue, T, Nishio, N, Hamanaka, S, Shimomura, T, Harada, S, Suzuki, S, Kohzuma, T, Shidara, S, Iwasaki, H, Kai, Y. | | Deposit date: | 1998-08-17 | | Release date: | 1999-08-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure determinations of oxidized and reduced pseudoazurins from Achromobacter cycloclastes. Concerted movement of copper site in redox forms with the rearrangement of hydrogen bond at a remote histidine.
J.Biol.Chem., 274, 1999
|
|
1ISR
 
 | | Crystal Structure of Metabotropic Glutamate Receptor Subtype 1 Complexed with Glutamate and Gadolinium Ion | | Descriptor: | GADOLINIUM ATOM, GLUTAMIC ACID, Metabotropic Glutamate Receptor subtype 1 | | Authors: | Tsuchiya, D, Kunishima, N, Kamiya, N, Jingami, H, Morikawa, K. | | Deposit date: | 2001-12-21 | | Release date: | 2002-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural views of the ligand-binding cores of a metabotropic glutamate receptor complexed with an antagonist and both glutamate and Gd3+.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1ARX
 
 | | CRYSTAL STRUCTURES OF CYANIDE-AND TRIIODIDE-BOUND FORMS OF ARTHROMYCES RAMOSUS PEROXIDASE AT DIFFERENT PH VALUES. PERTURBATIONS OF ACTIVE SITE RESIDUES AND THEIR IMPLICATION IN ENZYME CATALYSIS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Fukuyama, K, Kunishima, N, Amada, F. | | Deposit date: | 1995-04-25 | | Release date: | 1996-01-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of cyanide- and triiodide-bound forms of Arthromyces ramosus peroxidase at different pH values. Perturbations of active site residues and their implication in enzyme catalysis.
J.Biol.Chem., 270, 1995
|
|
1ARY
 
 | | CRYSTAL STRUCTURES OF CYANIDE-AND TRIIODIDE-BOUND FORMS OF ARTHROMYCES RAMOSUS PEROXIDASE AT DIFFERENT PH VALUES. PERTURBATIONS OF ACTIVE SITE RESIDUES AND THEIR IMPLICATION IN ENZYME CATALYSIS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Fukuyama, K, Kunishima, N, Amada, F. | | Deposit date: | 1995-04-25 | | Release date: | 1996-01-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of cyanide- and triiodide-bound forms of Arthromyces ramosus peroxidase at different pH values. Perturbations of active site residues and their implication in enzyme catalysis.
J.Biol.Chem., 270, 1995
|
|
1ISS
 
 | | Crystal Structure of Metabotropic Glutamate Receptor Subtype 1 Complexed with an antagonist | | Descriptor: | (S)-(ALPHA)-METHYL-4-CARBOXYPHENYLGLYCINE, Metabotropic Glutamate Receptor subtype 1 | | Authors: | Tsuchiya, D, Kunishima, N, Kamiya, N, Jingami, H, Morikawa, K. | | Deposit date: | 2001-12-21 | | Release date: | 2002-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural views of the ligand-binding cores of a metabotropic glutamate receptor complexed with an antagonist and both glutamate and Gd3+.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1J3B
 
 | | Crystal structure of ATP-dependent phosphoenolpyruvate carboxykinase from Thermus thermophilus HB8 | | Descriptor: | ATP-dependent phosphoenolpyruvate carboxykinase, CALCIUM ION, GLYCEROL, ... | | Authors: | Sugahara, M, Miyano, M, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-01-21 | | Release date: | 2003-02-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of ATP-dependent phosphoenolpyruvate carboxykinase from Thermus thermophilus HB8 showing the structural basis of induced fit and thermostability.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2ZSJ
 
 | |
3ADO
 
 | |
1J2W
 
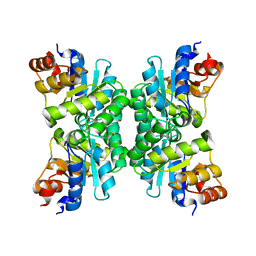 | | Tetrameric Structure of aldolase from Thermus thermophilus HB8 | | Descriptor: | Aldolase protein | | Authors: | Lokanath, N.K, Shiromizu, I, Miyano, M, Yokoyama, S, Kuramitsu, S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-01-14 | | Release date: | 2003-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of aldolase from Thermus thermophilus HB8 showing the contribution of oligomeric state to thermostability.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1AF3
 
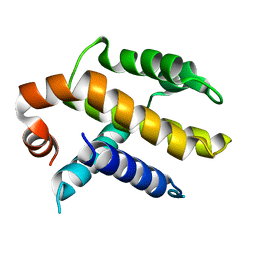 | | RAT BCL-XL AN APOPTOSIS INHIBITORY PROTEIN | | Descriptor: | APOPTOSIS REGULATOR BCL-X | | Authors: | Aritomi, M, Kunishima, N, Inohara, N, Ishibashi, Y, Ohta, S, Morikawa, K. | | Deposit date: | 1997-03-21 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of rat Bcl-xL. Implications for the function of the Bcl-2 protein family.
J.Biol.Chem., 272, 1997
|
|
2CUO
 
 | | Collagen model peptide (PRO-PRO-GLY)9 | | Descriptor: | COLLAGEN MODEL PEPTIDE (PRO-PRO-GLY)9 | | Authors: | Hongo, C, Noguchi, K, Okuyama, K, Tanaka, Y, Nishino, N. | | Deposit date: | 2005-05-27 | | Release date: | 2005-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Repetitive interactions observed in the crystal structure of a collagen-model peptide, [(Pro-Pro-Gly)9]3
J.Biochem.(Tokyo), 138, 2005
|
|
2OWE
 
 | | Crystal structure of TTHB049 from Thermus thermophilus HB8 | | Descriptor: | Alpha-ribazole-5'-phosphate phosphatase | | Authors: | Sugahara, M, Taketa, M, Kageyama, Y, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-16 | | Release date: | 2007-08-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of TTHB049 from Thermus thermophilus HB8
To be Published
|
|
2OWK
 
 | | Crystal structure of PH0725 from Pyrococcus horikoshii OT3 | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, diphthine synthase | | Authors: | Sugahara, M, Matsuura, Y, Ono, N, Nakamoto, T, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-16 | | Release date: | 2007-08-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of PH0725 from Pyrococcus horikoshii OT3
To be Published
|
|
2OWG
 
 | | Crystal structure of PH0725 from Pyrococcus horikoshii OT3 | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, diphthine synthase | | Authors: | Sugahara, M, Matsuura, Y, Tanaka, Y, Nakamoto, T, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-16 | | Release date: | 2007-08-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of PH0725 from Pyrococcus horikoshii OT3
To be Published
|
|
2OWU
 
 | | Crystal structure of PH0725 from Pyrococcus horikoshii OT3 | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, diphthine synthase | | Authors: | Sugahara, M, Taketa, M, Kageyama, Y, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-17 | | Release date: | 2007-08-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of PH0725 from Pyrococcus horikoshii OT3
To be Published
|
|
2P5C
 
 | | Crystal structure of PH0725 from Pyrococcus horikoshii OT3 | | Descriptor: | GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, diphthine synthase | | Authors: | Yamamoto, H, Taketa, M, Ono, N, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-15 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of PH0725 from Pyrococcus horikoshii OT3
To be Published
|
|
2P6O
 
 | | Crystal structure of TTHB049 from Thermus thermophilus HB8 | | Descriptor: | Alpha-ribazole-5'-phosphate phosphatase | | Authors: | Yamamoto, H, Taketa, M, Tanaka, Y, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-19 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of TTHB049 from Thermus thermophilus HB8
To be Published
|
|
