6K38
 
 | |
3CBF
 
 | | Crystal structure of LysN, alpha-aminoadipate aminotransferase, from Thermus thermophilus HB27 | | Descriptor: | (2S)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]hexanedioic acid, Alpha-aminodipate aminotransferase | | Authors: | Tomita, T, Miyazaki, T, Miyagawa, T, Fushinobu, S, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2008-02-21 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Mechanism for multiple-substrates recognition of alpha-aminoadipate aminotransferase from Thermus thermophilus
Proteins, 2008
|
|
2Z1Y
 
 | | Crystal structure of LysN, alpha-aminoadipate aminotransferase (complexed with N-(5'-phosphopyridoxyl)-L-leucine), from Thermus thermophilus HB27 | | Descriptor: | Alpha-aminodipate aminotransferase, LEUCINE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tomita, T, Miyazaki, T, Miyagawa, T, Fushinobu, S, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2007-05-16 | | Release date: | 2008-06-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanism of broad substrate specificity of alpha-aminoadipate aminotransferase from Thermus thermophilus
To be Published
|
|
6K37
 
 | | Crystal structure of BioU (K124A) from Synechocystis sp.PCC6803 in complex with NAD+ and the analog of reaction intermediate, 3-(1-aminoethyl)-nonanedioic acid | | Descriptor: | (3R)-3-[(1R)-1-azanylethyl]nonanedioic acid, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Slr0355 protein | | Authors: | Sakaki, K, Tomita, T, Nishiyama, M. | | Deposit date: | 2019-05-16 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A suicide enzyme catalyzes multiple reactions for biotin biosynthesis in cyanobacteria.
Nat.Chem.Biol., 16, 2020
|
|
3VPB
 
 | | ArgX from Sulfolobus tokodaii complexed with LysW/Glu/ADP/Mg/Zn/Sulfate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Alpha-aminoadipate carrier protein lysW, GLUTAMIC ACID, ... | | Authors: | Tomita, T, Ouchi, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2012-02-29 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Lysine and arginine biosyntheses mediated by a common carrier protein in Sulfolobus
Nat.Chem.Biol., 9, 2013
|
|
3A9I
 
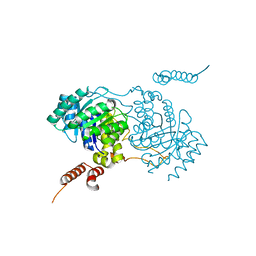 | | Crystal structure of homocitrate synthase from Thermus thermophilus complexed with Lys | | Descriptor: | COBALT (II) ION, Homocitrate synthase, LYSINE | | Authors: | Okada, T, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2009-10-28 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of substrate recognition and insight into feedback inhibition of homocitrate synthase from Thermus thermophilus
J.Biol.Chem., 285, 2010
|
|
2YR2
 
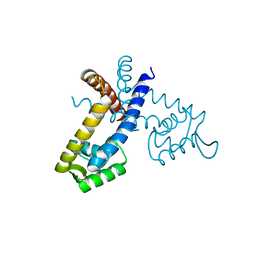 | |
3SO2
 
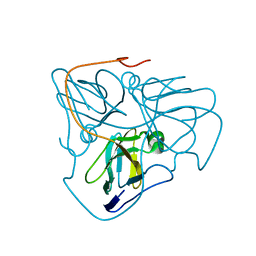 | | Chlorella dUTPase | | Descriptor: | Putative uncharacterized protein | | Authors: | Badalucco, L, Poudel, I, Natarajan, c, Yamanishi, M, Moriyama, H. | | Deposit date: | 2011-06-29 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6428 Å) | | Cite: | Crystallization of Chlorella deoxyuridine triphosphatase.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2Z9Z
 
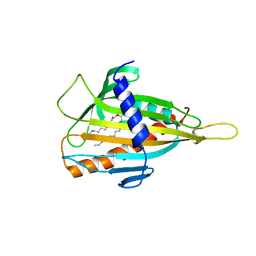 | | Crystal structure of CERT START domain(N504A mutant), in complex with C10-diacylglycerol | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl didecanoate, Lipid-transfer protein CERT | | Authors: | Kudo, N, Kumagai, K, Wakatsuki, S, Nishijima, M, Hanada, K, Kato, R. | | Deposit date: | 2007-09-26 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2RSG
 
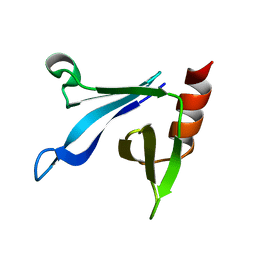 | | Solution structure of the CERT PH domain | | Descriptor: | Collagen type IV alpha-3-binding protein | | Authors: | Sugiki, T, Takeuchi, K, Tokunaga, Y, Kumagai, K, Kawano, M, Nishijima, M, Hanada, K, Takahashi, H, Shimada, I. | | Deposit date: | 2012-02-25 | | Release date: | 2012-08-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the Golgi association by the pleckstrin homology domain of the ceramide trafficking protein (CERT)
J.Biol.Chem., 287, 2012
|
|
3AUA
 
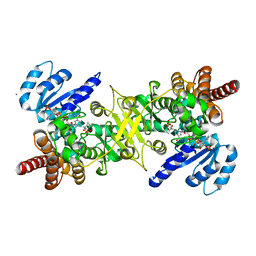 | | Crystal structure of the quaternary complex-2 of an isomerase | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[ethanoyl(hydroxy)amino]propylphosphonic acid, CALCIUM ION, ... | | Authors: | Umeda, T, Tanaka, N, Kusakabe, Y, Nakanishi, M, Kitade, Y, Nakamura, K.T. | | Deposit date: | 2011-02-01 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular basis of fosmidomycin's action on the human malaria parasite Plasmodium falciparum
Sci Rep, 1, 2011
|
|
3VOR
 
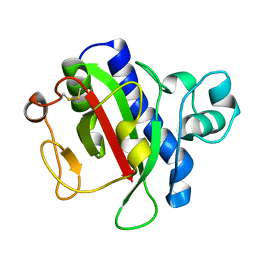 | | Crystal Structure Analysis of the CofA | | Descriptor: | CFA/III pilin | | Authors: | Fukakusa, S, Kawahara, K, Nakamura, S, Iwasita, T, Baba, S, Nishimura, M, Kobayashi, Y, Honda, T, Iida, T, Taniguchi, T, Ohkubo, T. | | Deposit date: | 2012-02-06 | | Release date: | 2012-09-26 | | Last modified: | 2013-07-31 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Structure of the CFA/III major pilin subunit CofA from human enterotoxigenic Escherichia coli determined at 0.90 A resolution by sulfur-SAD phasing
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3AU8
 
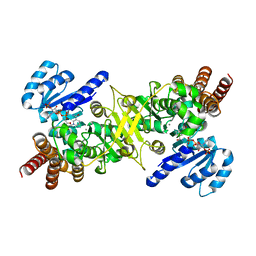 | | Crystal structure of the ternary complex of an isomerase | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, MANGANESE (II) ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Umeda, T, Tanaka, N, Kusakabe, Y, Nakanishi, M, Kitade, Y, Nakamura, K.T. | | Deposit date: | 2011-02-01 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Molecular basis of fosmidomycin's action on the human malaria parasite Plasmodium falciparum
Sci Rep, 1, 2011
|
|
3AU9
 
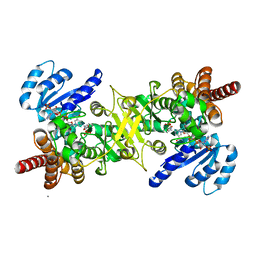 | | Crystal structure of the quaternary complex-1 of an isomerase | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, CALCIUM ION, ... | | Authors: | Umeda, T, Tanaka, N, Kusakabe, Y, Nakanishi, M, Kitade, Y, Nakamura, K.T. | | Deposit date: | 2011-02-01 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of fosmidomycin's action on the human malaria parasite Plasmodium falciparum
Sci Rep, 1, 2011
|
|
2ZTK
 
 | | Crystal structure of homocitrate synthase from Thermus thermophilus complexed with homocitrate | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, COPPER (II) ION, Homocitrate synthase | | Authors: | Okada, T, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2008-10-06 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Mechanism of substrate recognition and insight into feedback inhibition of homocitrate synthase from Thermus thermophilus
J.Biol.Chem., 285, 2010
|
|
2Z9Y
 
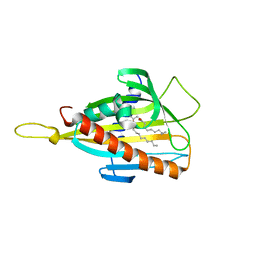 | | Crystal structure of CERT START domain in complex with C10-diacylglycerol | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl didecanoate, Lipid-transfer protein CERT | | Authors: | Kudo, N, Kumagai, K, Wakatsuki, S, Nishijima, M, Hanada, K, Kato, R. | | Deposit date: | 2007-09-26 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2ZTJ
 
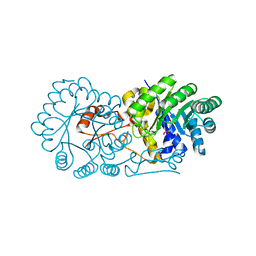 | | Crystal structure of homocitrate synthase from Thermus thermophilus complexed with alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, COPPER (II) ION, Homocitrate synthase | | Authors: | Okada, T, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2008-10-06 | | Release date: | 2009-10-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of substrate recognition and insight into feedback inhibition of homocitrate synthase from Thermus thermophilus
J.Biol.Chem., 285, 2010
|
|
2ZYF
 
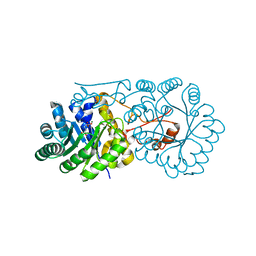 | | Crystal structure of homocitrate synthase from Thermus thermophilus complexed with magnesuim ion and alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, Homocitrate synthase, MAGNESIUM ION | | Authors: | Okada, T, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2009-01-20 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mechanism of substrate recognition and insight into feedback inhibition of homocitrate synthase from thermus thermophilus
J.Biol.Chem., 2009
|
|
2ZYJ
 
 | | Crystal structure of LysN, alpha-aminoadipate aminotransferase (complexed with N-(5'-phosphopyridoxyl)-L-glutamate), from Thermus thermophilus HB27 | | Descriptor: | Alpha-aminodipate aminotransferase, N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-glutamic acid | | Authors: | Ouchi, T, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2009-01-26 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Dual roles of a conserved pair, Arg23 and Ser20, in recognition of multiple substrates in alpha-aminoadipate aminotransferase from Thermus thermophilus.
Biochem.Biophys.Res.Commun., 388, 2009
|
|
1UC8
 
 | | Crystal structure of a lysine biosynthesis enzyme, Lysx, from thermus thermophilus HB8 | | Descriptor: | lysine biosynthesis enzyme | | Authors: | Sakai, H, Vassylyeva, M.N, Matsuura, T, Sekine, S, Nishiyama, M, Terada, T, Shirouzu, M, Kuramitsu, S, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-04-09 | | Release date: | 2003-09-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Lysine Biosynthesis Enzyme, LysX, from Thermus thermophilus HB8
J.Mol.Biol., 332, 2003
|
|
3VV5
 
 | | Crystal structure of TTC0807 complexed with (S)-2-aminoethyl-L-cysteine (AEC) | | Descriptor: | Amino acid ABC transporter, binding protein, L-THIALYSINE, ... | | Authors: | Tomita, T, Kanemaru, Y, Hasebe, F, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2012-07-16 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Two ATP-Binding Cassette Transporters Involved in (S)-2-Aminoethyl-Cysteine Uptake in Thermus thermophilus
J.Bacteriol., 195, 2013
|
|
3VVD
 
 | | Crystal structure of TTC0807 complexed with Ornithine | | Descriptor: | Amino acid ABC transporter, binding protein, L-ornithine | | Authors: | Tomita, T, Kanemaru, Y, Hasebe, F, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2012-07-21 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Two ATP-Binding Cassette Transporters Involved in (S)-2-Aminoethyl-Cysteine Uptake in Thermus thermophilus
J.Bacteriol., 195, 2013
|
|
3VVE
 
 | | Crystal structure of TTC0807 complexed with Lysine | | Descriptor: | Amino acid ABC transporter, binding protein, LYSINE, ... | | Authors: | Tomita, T, Kanemaru, Y, Hasebe, F, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2012-07-21 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two ATP-Binding Cassette Transporters Involved in (S)-2-Aminoethyl-Cysteine Uptake in Thermus thermophilus
J.Bacteriol., 195, 2013
|
|
3VPC
 
 | | ArgX from Sulfolobus tokodaii complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Putative acetylornithine deacetylase | | Authors: | Tomita, T, Horie, A, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2012-02-29 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Lysine and arginine biosyntheses mediated by a common carrier protein in Sulfolobus.
Nat.Chem.Biol., 9, 2013
|
|
3VVF
 
 | | Crystal structure of TTC0807 complexed with Arginine | | Descriptor: | ARGININE, Amino acid ABC transporter, binding protein, ... | | Authors: | Tomita, T, Kanemaru, Y, Hasebe, F, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2012-07-21 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Two ATP-Binding Cassette Transporters Involved in (S)-2-Aminoethyl-Cysteine Uptake in Thermus thermophilus
J.Bacteriol., 195, 2013
|
|
