4V1Y
 
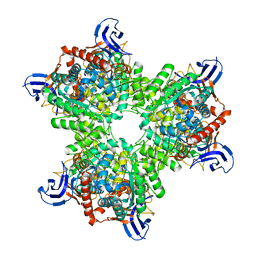 | | The structure of the hexameric atrazine chlorohydrolase, AtzA | | Descriptor: | 1,2-ETHANEDIOL, ATRAZINE CHLOROHYDROLASE, CHLORIDE ION, ... | | Authors: | Peat, T.S, Newman, J, Balotra, S, Lucent, D, Warden, A.C, Scott, C. | | Deposit date: | 2014-10-04 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure of the Hexameric Atrazine Chlorohydrolase Atza.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4V1X
 
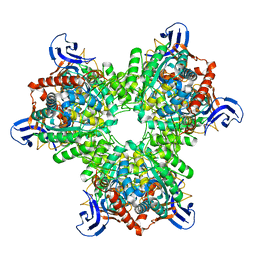 | | The structure of the hexameric atrazine chlorohydrolase, AtzA | | Descriptor: | ATRAZINE CHLOROHYDROLASE, DI(HYDROXYETHYL)ETHER, FE (III) ION | | Authors: | Peat, T.S, Newman, J, Balotra, S, Lucent, D, Warden, A.C, Scott, C. | | Deposit date: | 2014-10-04 | | Release date: | 2015-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of the Hexameric Atrazine Chlorohydrolase Atza.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3ZSO
 
 | | Small molecule inhibitors of the LEDGF site of HIV type 1 integrase identified by fragment screening and structure based design | | Descriptor: | 5-{[(2-{[bis(4-methoxyphenyl)methyl]carbamoyl}benzyl)(prop-2-en-1-yl)amino]methyl}-1,3-benzodioxole-4-carboxylic acid, ACETIC ACID, INTEGRASE, ... | | Authors: | Peat, T.S, Newman, J, Rhodes, D.I, Deadman, J.J, Vandergraaff, N, Le, G, Jones, E.D, Smith, J.A, Coates, J.A.V, Thienthong, N, Dolezal, O, Ryan, J.H, Savage, G.P, Francis, C.L. | | Deposit date: | 2011-06-30 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Small Molecule Inhibitors of the Ledgf Site of Human Immunodeficiency Virus Integrase Identified by Fragment Screening and Structure Based Design.
Plos One, 7, 2012
|
|
7MY1
 
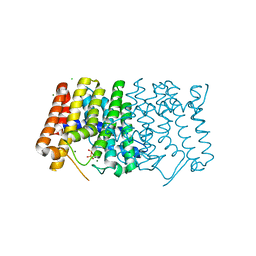 | | Sy-CrtE structure with IPP, N-term His-tag | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, CHLORIDE ION, Geranylgeranyl pyrophosphate synthase, ... | | Authors: | Peat, T.S, Newman, J. | | Deposit date: | 2021-05-19 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Molecular characterization of cyanobacterial short-chain prenyltransferases and discovery of a novel GGPP phosphatase.
Febs J., 289, 2022
|
|
7MXZ
 
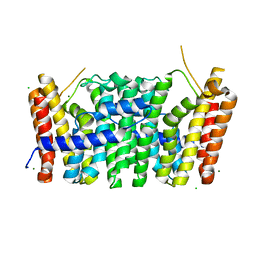 | | Sy-CrtE apo structure | | Descriptor: | CHLORIDE ION, Geranylgeranyl pyrophosphate synthase, MAGNESIUM ION | | Authors: | Peat, T.S, Newman, J. | | Deposit date: | 2021-05-19 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Molecular characterization of cyanobacterial short-chain prenyltransferases and discovery of a novel GGPP phosphatase.
Febs J., 289, 2022
|
|
7MY7
 
 | | Se-CrtE N-term His-tag structure | | Descriptor: | Farnesyl-diphosphate synthase | | Authors: | Peat, T.S, Newman, J. | | Deposit date: | 2021-05-20 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Molecular characterization of cyanobacterial short-chain prenyltransferases and discovery of a novel GGPP phosphatase.
Febs J., 289, 2022
|
|
7N9J
 
 | | Crystal structure of H2DB in complex with HSF2 melanoma neoantigen | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Patskovsky, Y, Finnigan, J, Patskovska, L, Newman, J, Bhardwaj, N, Krogsgaard, M. | | Deposit date: | 2021-06-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure of the complex between H2DB and melanoma HSF2 neoantigen YGFRNVVHI
To be Published
|
|
7NA5
 
 | | Structure of the H2DB-TCR ternary complex with HSF2 melanoma neoantigen | | Descriptor: | 47BE7 TCR alpha chain, 47BE7 TCR beta chain, Beta-2-microglobulin, ... | | Authors: | Patskovsky, Y, Finnigan, J, Patskovska, L, Newman, J, Bhardwaj, N, Krogsgaard, M. | | Deposit date: | 2021-06-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the TCR-H2DB ternary complex with melanoma HSF2 neoantigen YGFRNVVHI
To be Published
|
|
7MY6
 
 | | Se-CrtE C-term His-tag with IPP added | | Descriptor: | 1,2-ETHANEDIOL, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Peat, T.S, Newman, J. | | Deposit date: | 2021-05-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Molecular characterization of cyanobacterial short-chain prenyltransferases and discovery of a novel GGPP phosphatase.
Febs J., 289, 2022
|
|
6NWM
 
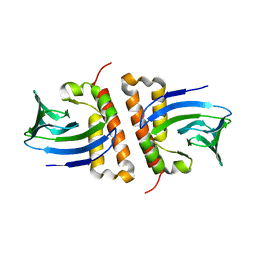 | |
6NWO
 
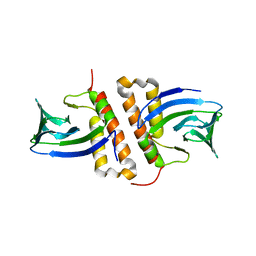 | | Structures of the transcriptional regulator BgaR, a lactose sensor. | | Descriptor: | CHLORIDE ION, GLYCEROL, Transcriptional regulator BgaR, ... | | Authors: | Peat, T.S, Newman, J. | | Deposit date: | 2019-02-06 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structures of the transcriptional regulator BgaR, a lactose sensor.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6NX3
 
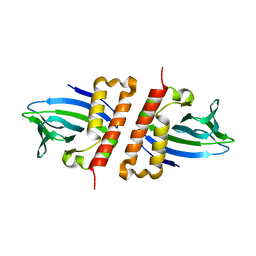 | |
6NWH
 
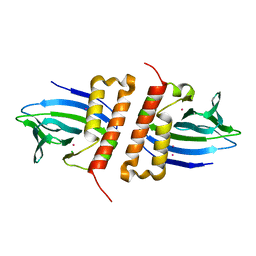 | | Structures of the transcriptional regulator BgaR, a lactose sensor. | | Descriptor: | CHLORIDE ION, MERCURY (II) ION, Transcriptional regulator BgaR, ... | | Authors: | Peat, T.S, Newman, J. | | Deposit date: | 2019-02-06 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of the transcriptional regulator BgaR, a lactose sensor.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6NWJ
 
 | |
8TFO
 
 | | Structure of MKvar | | Descriptor: | (R)-MEVALONATE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Peat, T.S, Newman, J, Esquirol, L, Nebl, T, Scott, C, Vickers, C, Sainsbury, F. | | Deposit date: | 2023-07-11 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of novel mevalonate kinases from the tardigrade Ramazzottius varieornatus and the psychrophilic archaeon Methanococcoides burtonii.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8TEB
 
 | | Structure of MKbur | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Peat, T.S, Newman, J, Esquirol, L, Nebl, T, Scott, C, Vickers, C, Sainsbury, F. | | Deposit date: | 2023-07-06 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of novel mevalonate kinases from the tardigrade Ramazzottius varieornatus and the psychrophilic archaeon Methanococcoides burtonii.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
3PN1
 
 | | Novel Bacterial NAD+-dependent DNA Ligase Inhibitors with Broad Spectrum Potency and Antibacterial Efficacy In Vivo | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 2-(butylsulfanyl)adenosine, DNA ligase | | Authors: | Mills, S, Eakin, A, Buurman, E, Newman, J, Gao, N, Huynh, H, Johnson, K, Lahiri, S, Shapiro, A, Walkup, G, Wei, Y, Stokes, S. | | Deposit date: | 2010-11-18 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel Bacterial NAD+-Dependent DNA Ligase Inhibitors with Broad-Spectrum Activity and Antibacterial Efficacy In Vivo.
Antimicrob.Agents Chemother., 55, 2011
|
|
1BKB
 
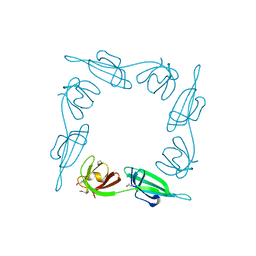 | | INITIATION FACTOR 5A FROM ARCHEBACTERIUM PYROBACULUM AEROPHILUM | | Descriptor: | TRANSLATION INITIATION FACTOR 5A | | Authors: | Peat, T.S, Newman, J, Waldo, G.S, Berendzen, J, Terwilliger, T.C. | | Deposit date: | 1998-07-05 | | Release date: | 1998-11-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of translation initiation factor 5A from Pyrobaculum aerophilum at 1.75 A resolution.
Structure, 6, 1998
|
|
4UHN
 
 | | Characterization of a Novel Transaminase from Pseudomonas sp. Strain AAC | | Descriptor: | ACETIC ACID, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wilding, M, Peat, T.S, Newman, J, Scott, C. | | Deposit date: | 2015-03-25 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | A Beta-Alanine Catabolism Pathway Containing a Highly Promiscuous Omega-Transaminase in the 12-Aminododecanate-Degrading Pseudomonas Sp. Strain Aac.
Appl.Environ.Microbiol., 82, 2016
|
|
4UHM
 
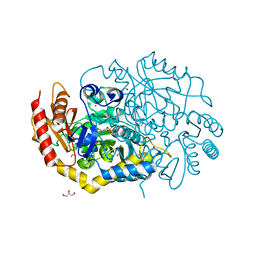 | | Characterization of a Novel Transaminase from Pseudomonas sp. Strain AAC | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, ETHANOL, ... | | Authors: | Wilding, M, Peat, T.S, Newman, J, Scott, C. | | Deposit date: | 2015-03-25 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | A Beta-Alanine Catabolism Pathway Containing a Highly Promiscuous Omega-Transaminase in the 12-Aminododecanate-Degrading Pseudomonas Sp. Strain Aac.
Appl.Environ.Microbiol., 82, 2016
|
|
4UHO
 
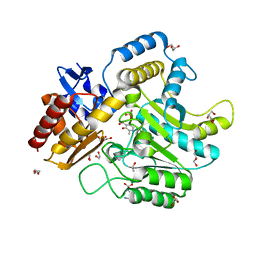 | | Characterization of a Novel Transaminase from Pseudomonas sp. Strain AAC | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ETHANOL, ... | | Authors: | Wilding, M, Peat, T.S, Newman, J, Scott, C. | | Deposit date: | 2015-03-25 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | A Beta-Alanine Catabolism Pathway Containing a Highly Promiscuous Omega-Transaminase in the 12-Aminododecanate-Degrading Pseudomonas Sp. Strain Aac.
Appl.Environ.Microbiol., 82, 2016
|
|
7K09
 
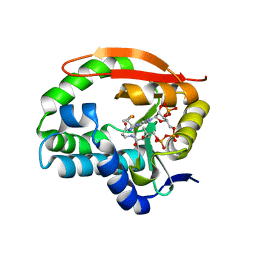 | | Puromycin N-acetyltransferase in complex with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, Puromycin N-acetyltransferase | | Authors: | Caputo, A.T, Newman, J, Adams, T.E, Peat, T.S. | | Deposit date: | 2020-09-03 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.313 Å) | | Cite: | Structure-guided selection of puromycin N-acetyltransferase mutants with enhanced selection stringency for deriving mammalian cell lines expressing recombinant proteins.
Sci Rep, 11, 2021
|
|
7K0A
 
 | | Puromycin N-acetyltransferase in complex with acetylated puromycin and CoA | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Peat, T.S, Caputo, A.T, Newman, J, Adams, T.E. | | Deposit date: | 2020-09-03 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided selection of puromycin N-acetyltransferase mutants with enhanced selection stringency for deriving mammalian cell lines expressing recombinant proteins.
Sci Rep, 11, 2021
|
|
7K7V
 
 | | The X-ray crystal structure of SSR4, an S. pombe chromatin remodelling protein: iodide derivative | | Descriptor: | CHLORIDE ION, GLYCEROL, IODIDE ION, ... | | Authors: | Peat, T.S, Newman, J. | | Deposit date: | 2020-09-24 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | The X-ray crystal structure of the N-terminal domain of Ssr4, a Schizosaccharomyces pombe chromatin-remodelling protein.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
1CV2
 
 | | Hydrolytic haloalkane dehalogenase linb from sphingomonas paucimobilis UT26 AT 1.6 A resolution | | Descriptor: | HALOALKANE DEHALOGENASE | | Authors: | Marek, J, Vevodova, J, Damborsky, J, Smatanova, I, Svensson, L.A, Newman, J, Nagata, Y, Takagi, M. | | Deposit date: | 1999-08-22 | | Release date: | 2000-09-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of the haloalkane dehalogenase from Sphingomonas paucimobilis UT26.
Biochemistry, 39, 2000
|
|
