5KR3
 
 | | Directed Evolution of Transaminases By Ancestral Reconstruction. Using Old Proteins for New Chemistries | | Descriptor: | 4-aminobutyrate transaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Wilding, M, Newman, J, Peat, T.S, Scott, C. | | Deposit date: | 2016-07-06 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Reverse engineering: transaminase biocatalyst development using ancestral sequence reconstruction
Green Chemistry, 19, 2017
|
|
5KQW
 
 | | Directed Evolution of Transaminases By Ancestral Reconstruction. Using Old Proteins for New Chemistries | | Descriptor: | 1,2-ETHANEDIOL, 4-aminobutyrate transaminase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wilding, M, Newman, J, Peat, T.S, Scott, C. | | Deposit date: | 2016-07-06 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Reverse engineering: transaminase biocatalyst development using ancestral sequence reconstruction
Green Chemistry, 19, 2017
|
|
5KR5
 
 | | Directed Evolution of Transaminases By Ancestral Reconstruction. Using Old Proteins for New Chemistries | | Descriptor: | 4-aminobutyrate transaminase, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wilding, M, Newman, J, Peat, T.S, Scott, C. | | Deposit date: | 2016-07-07 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Reverse engineering: transaminase biocatalyst development using ancestral sequence reconstruction
Green Chemistry, 19, 2017
|
|
6G0Q
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with an acetylated GATA1 peptide (K312ac/K315ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, Erythroid transcription factor | | Authors: | Filippakopoulos, P, Picaud, S, Newman, J, Sorrell, F, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | Deposit date: | 2018-03-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
5AKQ
 
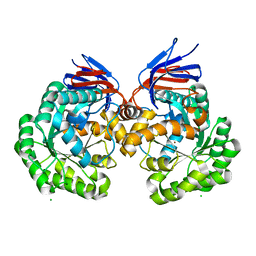 | | X-ray structure and mutagenesis studies of the N-isopropylammelide isopropylaminohydrolase, AtzC | | Descriptor: | CHLORIDE ION, N-ISOPROPYLAMMELIDE ISOPROPYL AMIDOHYDROLASE, ZINC ION | | Authors: | Balotra, S, Warden, A.C, Newman, J, Briggs, L.J, Scott, C, Peat, T.S. | | Deposit date: | 2015-03-05 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-Ray Structure and Mutagenesis Studies of the N-Isopropylammelide Isopropylaminohydrolase, Atzc
Plos One, 1, 2015
|
|
5A25
 
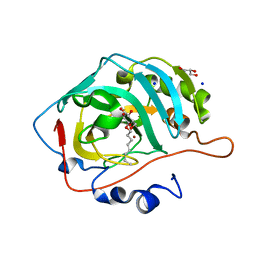 | | Rational engineering of a mesophilic carbonic anhydrase to an extreme halotolerant biocatalyst | | Descriptor: | CARBONIC ANHYDRASE 2, GLYCEROL, SODIUM ION, ... | | Authors: | Warden, A, Newman, J, Peat, T.S, Seabrook, S, Williams, M, Dojchinov, G, Haritos, V. | | Deposit date: | 2015-05-12 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational Engineering of a Mesohalophilic Carbonic Anhydrase to an Extreme Halotolerant Biocatalyst.
Nat.Commun., 6, 2015
|
|
5BK6
 
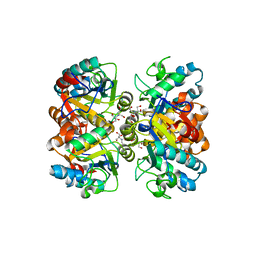 | | Structural and biochemical characterization of a non-canonical biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Putative amidase | | Authors: | Peat, T.S, Esquirol, L, Newman, J, Scott, C. | | Deposit date: | 2017-09-12 | | Release date: | 2018-02-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural and biochemical characterization of the biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841.
PLoS ONE, 13, 2018
|
|
7PSP
 
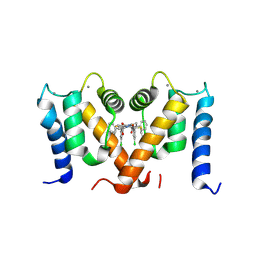 | | Crystal structure of S100A4 labeled with NU000846b. | | Descriptor: | (2R,4R)-1-(2-chloranylethanoyl)-N-(3-chlorophenyl)-4-phenyl-pyrrolidine-2-carboxamide, CALCIUM ION, Protein S100-A4 | | Authors: | Giroud, C, Szommer, T, Coxon, C, Monteiro, O, Christott, T, Bennett, J, Aitmakhanova, K, Raux, B, Newman, J, Elkins, J, Arruda Bezerra, G, Krojer, T, Koekemoer, L, Von Delft, F, Bountr, C, Brennan, P, Fedorov, O. | | Deposit date: | 2021-09-23 | | Release date: | 2022-10-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of S100A4 labeled with NU000846b.
To Be Published
|
|
6R7T
 
 | | Crystal Structure of human Melanoma-associated antigen B1 (MAGEB1) in complex with nanobody | | Descriptor: | Melanoma-associated antigen B1, anti MAGEB1 nanobody | | Authors: | Ye, M, Newman, J, Pike, A.C.W, Burgess-Brown, N, Cooper, C.D.O, Bountra, C, Arrowsmith, C, Edwards, A, Gileadi, O, von Delft, F. | | Deposit date: | 2019-03-29 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.682 Å) | | Cite: | Crystal Structure of Melanoma-associated antigen B1 (MAGEB1) in complex with nanobody
To Be Published
|
|
6AZN
 
 | | Structural and biochemical characterization of a non-canonical biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841 | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Putative amidase | | Authors: | Peat, T.S, Esquirol, L, Newman, J, Scott, C. | | Deposit date: | 2017-09-11 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and biochemical characterization of the biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841.
PLoS ONE, 13, 2018
|
|
6AZQ
 
 | | Structural and biochemical characterization of a non-canonical biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841 | | Descriptor: | CALCIUM ION, Putative amidase, dicarbonimidic diamide | | Authors: | Peat, T.S, Esquirol, L, Newman, J, Scott, C. | | Deposit date: | 2017-09-11 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural and biochemical characterization of the biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841.
PLoS ONE, 13, 2018
|
|
6BJT
 
 | | The structure of AtzH: a little known member of the atrazine breakdown pathway | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Peat, T.S, Newman, J, Scott, C, Esquirol, L. | | Deposit date: | 2017-11-07 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel decarboxylating amidohydrolase involved in avoiding metabolic dead ends during cyanuric acid catabolism in Pseudomonas sp. strain ADP.
PLoS ONE, 13, 2018
|
|
6BJU
 
 | |
8W0O
 
 | | GDH-105 crystal structure | | Descriptor: | CHLORIDE ION, Dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Peat, T.S, Newman, J. | | Deposit date: | 2024-02-13 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Enhancing the Imine Reductase Activity of a Promiscuous Glucose Dehydrogenase for Scalable Manufacturing of a Chiral Neprilysin Inhibitor Precursor
Acs Catalysis, 14, 2024
|
|
8W0N
 
 | | IRED crystal structure | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, IRED, ... | | Authors: | Peat, T.S, Newman, J. | | Deposit date: | 2024-02-13 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Enhancing the Imine Reductase Activity of a Promiscuous Glucose Dehydrogenase for Scalable Manufacturing of a Chiral Neprilysin Inhibitor Precursor
Acs Catalysis, 14, 2024
|
|
6D63
 
 | | The structure of AtzH: a little known member of the atrazine breakdown pathway | | Descriptor: | 3-oxopentanedioic acid, atzH | | Authors: | Peat, T.S, Newman, J, Scott, C, Esquirol, L. | | Deposit date: | 2018-04-19 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel decarboxylating amidohydrolase involved in avoiding metabolic dead ends during cyanuric acid catabolism in Pseudomonas sp. strain ADP.
PLoS ONE, 13, 2018
|
|
4BJO
 
 | | Nitrate in the active site of PTP1b is a putative mimetic of the transition state | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Kenny, P.W, Newman, J, Peat, T.S. | | Deposit date: | 2013-04-19 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Nitrate in the Active Site of Protein Tyrosine Phosphatase 1B is a Putative Mimetic of the Transition State.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4BVR
 
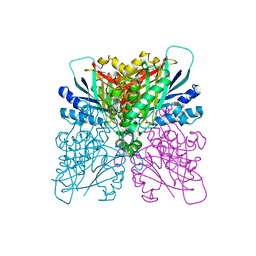 | | Cyanuric acid hydrolase: evolutionary innovation by structural concatenation. | | Descriptor: | 1,3,5-triazine-2,4,6-triol, CYANURIC ACID AMIDOHYDROLASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Peat, T.S, Balotra, S, Wilding, M, French, N.G, Briggs, L.J, Panjikar, S, Cowieson, N, Newman, J, Scott, C. | | Deposit date: | 2013-06-28 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Cyanuric Acid Hydrolase: Evolutionary Innovation by Structural Concatenation.
Mol.Microbiol., 88, 2013
|
|
4BVQ
 
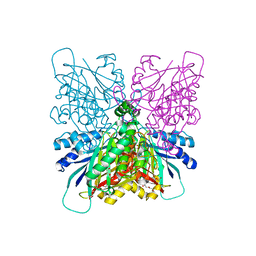 | | Cyanuric acid hydrolase: evolutionary innovation by structural concatenation. | | Descriptor: | CYANURIC ACID AMIDOHYDROLASE, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Peat, T.S, Balotra, S, Wilding, M, French, N.G, Briggs, L.J, Panjikar, S, Cowieson, N, Newman, J, Scott, C. | | Deposit date: | 2013-06-28 | | Release date: | 2013-07-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cyanuric Acid Hydrolase: Evolutionary Innovation by Structural Concatenation.
Mol.Microbiol., 88, 2013
|
|
4BVT
 
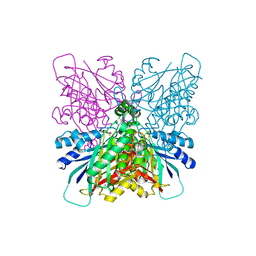 | | Cyanuric acid hydrolase: evolutionary innovation by structural concatenation | | Descriptor: | BARBITURIC ACID, CYANURIC ACID AMIDOHYDROLASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Peat, T.S, Balotra, S, Wilding, M, French, N.G, Briggs, L.J, Panjikar, S, Cowieson, N, Newman, J, Scott, C. | | Deposit date: | 2013-06-28 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Cyanuric Acid Hydrolase: Evolutionary Innovation by Structural Concatenation.
Mol.Microbiol., 88, 2013
|
|
4BVS
 
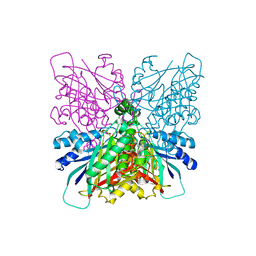 | | Cyanuric acid hydrolase: evolutionary innovation by structural concatenation. | | Descriptor: | 1,3,5-triazine-2,4,6-triamine, CYANURIC ACID AMIDOHYDROLASE, MAGNESIUM ION | | Authors: | Peat, T.S, Balotra, S, Wilding, M, French, N.G, Briggs, L.J, Panjikar, S, Cowieson, N, Newman, J, Scott, C. | | Deposit date: | 2013-06-28 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Cyanuric Acid Hydrolase: Evolutionary Innovation by Structural Concatenation.
Mol.Microbiol., 88, 2013
|
|
4CYY
 
 | | The structure of vanin-1: defining the link between metabolic disease, oxidative stress and inflammation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PANTETHEINASE | | Authors: | Boersma, Y.L, Newman, J, Adams, T.E, Sparrow, L, Cowieson, N, Lucent, D, Krippner, G, Bozaoglu, K, Peat, T.S. | | Deposit date: | 2014-04-16 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | The Structure of Vanin-1: A Key Enzyme Linking Metabolic Disease and Inflammation
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CYG
 
 | | The structure of vanin-1: defining the link between metabolic disease, oxidative stress and inflammation | | Descriptor: | (2R)-2,4-dihydroxy-N-[(3S)-3-hydroxy-4-phenylbutyl]-3,3-dimethylbutanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Boersma, Y.L, Newman, J, Adams, T.E, Sparrow, L, Cowieson, N, Lucent, D, Krippner, G, Bozaoglu, K, Peat, T.S. | | Deposit date: | 2014-04-11 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure of Vanin-1: A Key Enzyme Linking Metabolic Disease and Inflammation
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CQC
 
 | | The reaction mechanism of the N-isopropylammelide isopropylaminohydrolase AtzC: insights from structural and mutagenesis studies | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, N-ISOPROPYLAMMELIDE ISOPROPYL AMIDOHYDROLASE, ... | | Authors: | Balotra, S, Newman, J, French, N.G, Peat, T.S, Scott, C. | | Deposit date: | 2014-02-13 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-Ray Structure and Mutagenesis Studies of the N-Isopropylammelide Isopropylaminohydrolase, Atzc
Plos One, 1, 2015
|
|
4CNX
 
 | | Surface residue engineering of bovine carbonic anhydrase to an extreme halophilic enzyme for potential application in postcombustion CO2 capture | | Descriptor: | CARBONIC ANHYDRASE 2, DI(HYDROXYETHYL)ETHER, ZINC ION | | Authors: | Warden, A, Newman, J, Peat, T.S, Seabrook, S, Williams, M, Dojchinov, G, Haritos, V. | | Deposit date: | 2014-01-25 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Rational Engineering of a Mesohalophilic Carbonic Anhydrase to an Extreme Halotolerant Biocatalyst.
Nat.Commun., 6, 2015
|
|
