3HDV
 
 | |
3HEB
 
 | |
3HDC
 
 | |
1WUF
 
 | | Crystal structure of protein GI:16801725, member of Enolase superfamily from Listeria innocua Clip11262 | | Descriptor: | MAGNESIUM ION, hypothetical protein lin2664 | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-07 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3IN1
 
 | |
2IM5
 
 | | Crystal structure of Nicotinate phosphoribosyltransferase from Porphyromonas Gingivalis | | Descriptor: | Nicotinate phosphoribosyltransferase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-03 | | Release date: | 2006-10-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Nicotinate phosphoribosyltransferase from Porphyromonas gingivalis
To be Published
|
|
3IH0
 
 | | Crystal structure of an uncharacterized sugar kinase PH1459 from Pyrococcus horikoshii in complex with AMP-PNP | | Descriptor: | GLYCEROL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Uncharacterized sugar kinase PH1459 | | Authors: | Kumar, G, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-29 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an uncharacterized sugar kinase PH1459 from Pyrococcus horikoshii in complex with AMP-PNP
To be Published
|
|
3IA1
 
 | | Crystal structure of thio-disulfide isomerase from Thermus thermophilus | | Descriptor: | ACETATE ION, Thio-disulfide isomerase/thioredoxin | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-13 | | Release date: | 2009-07-21 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of thio-disulfide isomerase from Thermus thermophilus
To be Published
|
|
3IJ6
 
 | | CRYSTAL STRUCTURE OF AN UNCHARACTERIZED METAL-DEPENDENT HYDROLASE FROM Lactobacillus acidophilus | | Descriptor: | SODIUM ION, UNCHARACTERIZED METAL-DEPENDENT HYDROLASE, ZINC ION | | Authors: | Patskovsky, Y, Toro, R, Dickey, M, Chang, S, Sauder, J.M, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-03 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF AN UNCHARACTERIZED METAL-DEPENDENT HYDROLASE FROM Lactobacillus acidopphilus
To be Published
|
|
3IO1
 
 | | Crystal Structure of Aminobenzoyl-glutamate utilization protein from Klebsiella pneumoniae | | Descriptor: | Aminobenzoyl-glutamate utilization protein, SODIUM ION, YTTRIUM (III) ION | | Authors: | Kumaran, D, Baumann, K, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-13 | | Release date: | 2009-08-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Aminobenzoyl-glutamate utilization protein from Klebsiella pneumoniae
To be Published
|
|
3IOY
 
 | |
3IRV
 
 | | CRYSTAL STRUCTURE OF CYSTEINE HYDROLASE PSPPH_2384 FROM Pseudomonas syringae pv. phaseolicola 1448A | | Descriptor: | CYSTEINE HYDROLASE, GLYCEROL, PHOSPHATE ION | | Authors: | Patskovsky, Y, Malashkevich, V, Toro, R, Foti, R, Dickey, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-24 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | CRYSTAL STRUCTURE OF CYSTEINE HYDROLASE PSPPH_2384 FROM Pseudomonas syringae
To be Published
|
|
3IKH
 
 | |
2I3O
 
 | |
3ICJ
 
 | | Crystal structure of an uncharacterized metal-dependent hydrolase from pyrococcus furiosus | | Descriptor: | ZINC ION, uncharacterized metal-dependent hydrolase | | Authors: | Bonanno, J.B, Patskovsky, Y, Freeman, J, Bain, K.T, Hu, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-17 | | Release date: | 2009-07-28 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of an Uncharacterized Metal-Dependent Hydrolase from Pyrococcus Furiosus
To be Published
|
|
3ID9
 
 | |
3IE7
 
 | |
3IGH
 
 | | Crystal structure of an uncharacterized metal-dependent hydrolase from pyrococcus horikoshii ot3 | | Descriptor: | SULFATE ION, UNCHARACTERIZED METAL-DEPENDENT HYDROLASE | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-27 | | Release date: | 2009-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of an Uncharacterized Metal-Dependent Hydrolase from Pyrococcus Horikoshii
To be Published
|
|
3IWA
 
 | | Crystal structure of a FAD-dependent pyridine nucleotide-disulphide oxidoreductase from Desulfovibrio vulgaris | | Descriptor: | CALCIUM ION, FAD-dependent pyridine nucleotide-disulphide oxidoreductase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Iizuka, M, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-02 | | Release date: | 2009-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a FAD-dependent pyridine nucleotide-disulphide oxidoreductase from Desulfovibrio vulgaris
To be Published
|
|
3IPI
 
 | | Crystal Structure of a Geranyltranstransferase from the Methanosarcina mazei | | Descriptor: | Geranyltranstransferase, MALONIC ACID | | Authors: | Kumaran, D, Mohammed, M.B, Brown, A, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-17 | | Release date: | 2009-09-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of a Geranyltranstransferase from the Methanosarcina mazei
To be Published
|
|
3I83
 
 | | Crystal structure of 2-dehydropantoate 2-reductase from Methylococcus capsulatus | | Descriptor: | 2-dehydropantoate 2-reductase, ACETIC ACID | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Chang, S, Sampathkumar, P, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-09 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of 2-dehydropantoate 2-reductase from Methylococcus capsulatus
To be Published
|
|
3I9X
 
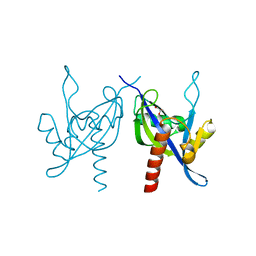 | | Crystal structure of a mutT/nudix family protein from Listeria innocua | | Descriptor: | GLYCEROL, mutT/nudix family protein | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Miller, S, Romero, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-13 | | Release date: | 2009-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a mutT/nudix family protein from Listeria innocua
To be Published
|
|
3IMH
 
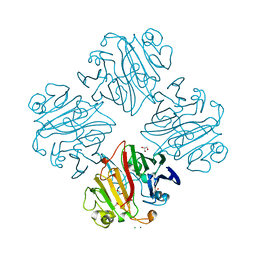 | | CRYSTAL STRUCTURE OF GALACTOSE 1-EPIMERASE FROM Lactobacillus acidophilus NCFM | | Descriptor: | CHLORIDE ION, GLYCEROL, Galactose-1-epimerase, ... | | Authors: | Patskovsky, Y, Toro, R, Dickey, M, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-10 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | CRYSTAL STRUCTURE OF GALACTOSE 1-EPIMERASE FROM Lactobacillus acidophilus
To be Published
|
|
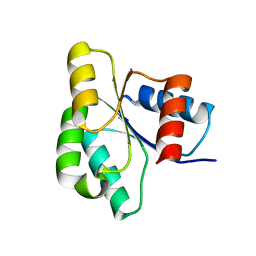
3ILH
 
 | |
2HAE
 
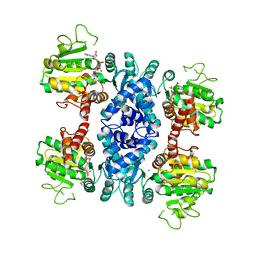 | |
