2NRQ
 
 | |
2O16
 
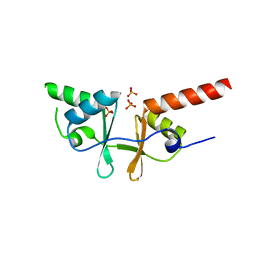 | | Crystal structure of a putative acetoin utilization protein (AcuB) from Vibrio cholerae | | Descriptor: | Acetoin utilization protein AcuB, putative, PHOSPHATE ION | | Authors: | Patskovsky, Y, Bonanno, J.B, Rutter, M, Bain, K.T, Powell, A, Slocombe, A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-28 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a putative acetoin utilization protein (AcuB) from Vibrio cholerae
To be Published
|
|
3KW2
 
 | |
3KZB
 
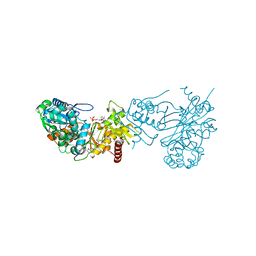 | |
3IVE
 
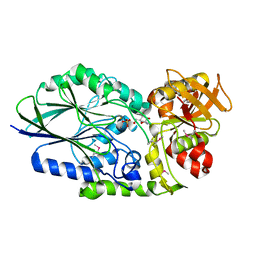 | | Putative 5'-Nucleotidase (c4898) from Escherichia Coli in complex with Cytidine | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, CHLORIDE ION, FE (III) ION, ... | | Authors: | Ramagopal, U.A, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-31 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Putative 5'-Nucleotidase (c4898) from Escherichia Coli in complex with Cytidine
To be Published
|
|
2NWU
 
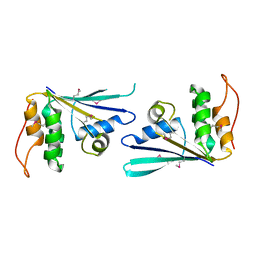 | |
3K9D
 
 | | CRYSTAL STRUCTURE OF PROBABLE ALDEHYDE DEHYDROGENASE FROM Listeria monocytogenes EGD-e | | Descriptor: | ALDEHYDE DEHYDROGENASE, CHLORIDE ION, GLYCEROL | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-15 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Aldehyde Dehydrogenase from Listeria Monocytogenes
To be Published
|
|
3HUR
 
 | | Crystal structure of alanine racemase from Oenococcus oeni | | Descriptor: | Alanine racemase, SULFATE ION | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Hu, S, Romero, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-06-15 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of alanine racemase from Oenococcus oeni
To be Published
|
|
2O34
 
 | | Crystal structure of protein DVU1097 from Desulfovibrio vulgaris Hildenborough, Pfam DUF375 | | Descriptor: | Hypothetical protein, SODIUM ION | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Schwinn, K.D, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Emtage, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-30 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the Hypothetical Protein from Desulfovibrio vulgaris Hildenborough
To be Published
|
|
3HMU
 
 | | Crystal structure of a class III aminotransferase from Silicibacter pomeroyi | | Descriptor: | Aminotransferase, class III, CHLORIDE ION, ... | | Authors: | Toro, R, Bonanno, J.B, Ramagopal, U, Freeman, J, Bain, K.T, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-29 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a class III aminotransferase from Silicibacter pomeroyi
To be Published
|
|
2OGF
 
 | | Crystal structure of protein MJ0408 from Methanococcus jannaschii, Pfam DUF372 | | Descriptor: | 8-OXOGUANINE, GLYCEROL, Hypothetical protein MJ0408, ... | | Authors: | Ramagopal, U.A, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-05 | | Release date: | 2007-01-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of hypothetical protein MJ0408 from Methanococcus jannaschii
To be Published
|
|
2OGK
 
 | |
3JU2
 
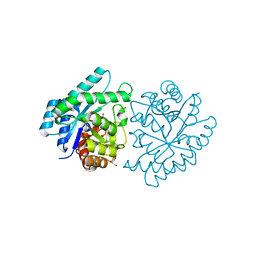 | | CRYSTAL STRUCTURE OF PROTEIN SMc04130 FROM Sinorhizobium meliloti 1021 | | Descriptor: | GLYCEROL, ZINC ION, uncharacterized protein SMc04130 | | Authors: | Patskovsky, Y, Foti, R, Ramagopal, U, Malashkevich, V, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-14 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURE OF PROTEIN SMc04130 FROM Sinorhizobium meliloti
To be Published
|
|
3JZV
 
 | |
2OOG
 
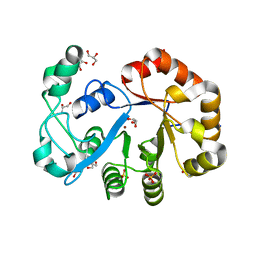 | | Crystal structure of glycerophosphoryl diester phosphodiesterase from Staphylococcus aureus | | Descriptor: | GLYCEROL, Glycerophosphoryl diester phosphodiesterase, SULFATE ION, ... | | Authors: | Patskovsky, Y, Fedorov, E, Toro, R, Sauder, J.M, Smith, D, Freeman, J, Maletic, M, Powell, A, Gheyi, T, Wasserman, S.R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-25 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Glycerophosphoryl Diester Phosphodiesterase from Staphylococcus Aureus
To be Published
|
|
3L0Q
 
 | |
3I4J
 
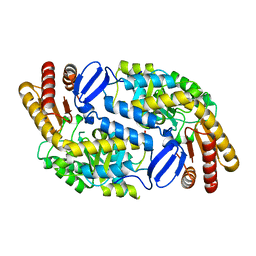 | | Crystal structure of Aminotransferase, class III from Deinococcus radiodurans | | Descriptor: | Aminotransferase, class III, SULFATE ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-01 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Aminotransferase, class III from Deinococcus radiodurans
To be Published
|
|
3HPA
 
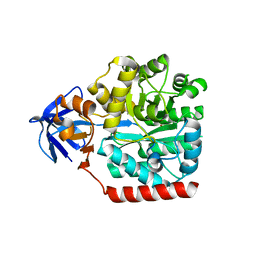 | | Crystal structure of an amidohydrolase gi:44264246 from an evironmental sample of sargasso sea | | Descriptor: | AMIDOHYDROLASE, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-06-03 | | Release date: | 2009-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The hunt for 8-oxoguanine deaminase.
J.Am.Chem.Soc., 132, 2010
|
|
2PHP
 
 | |
2PGE
 
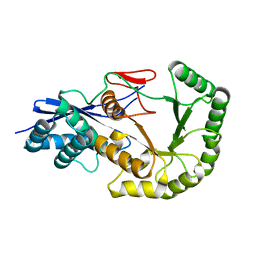 | |
3HZ6
 
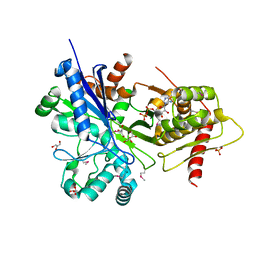 | |
3I6E
 
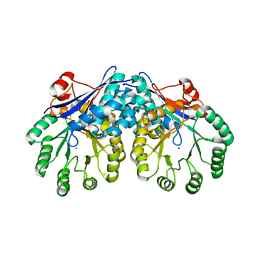 | | CRYSTAL STRUCTURE OF MUCONATE LACTONIZING ENZYME FROM Ruegeria pomeroyi. | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase I, SODIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-07 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of muconate lactonizing enzyme from Ruegeria pomeroyi.
To be Published
|
|
3I42
 
 | |
3LM4
 
 | | Crystal Structure of 2,3-Dihydroxy Biphenyl dioxygenase from Rhodococcus sp. (strain RHA1) | | Descriptor: | (2Z,4E)-2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOIC ACID, Catechol 2,3-dioxygenase, FE (III) ION, ... | | Authors: | Syed Ibrahim, B, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-01-29 | | Release date: | 2010-02-23 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of 2,3-Dihydroxy Biphenyl dioxygenase from Rhodococcus sp. (strain RHA1)
To be Published
|
|
2PAJ
 
 | |
