2IQI
 
 | | Crystal structure of protein XCC0632 from Xanthomonas campestris, Pfam DUF330 | | Descriptor: | Hypothetical protein XCC0632 | | Authors: | Bonanno, J.B, Gilmore, J, Bain, K.T, Mckenzie, C, Pelletier, L, Wasserman, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-13 | | Release date: | 2006-11-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of hypothetical protein XCC0632 from Xanthomonas campestris pv. campestris
To be Published
|
|
2HAF
 
 | | Crystal structure of a putative translation repressor from Vibrio cholerae | | Descriptor: | Putative translation repressor | | Authors: | Sugadev, R, Seetharaman, J, Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-06-12 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Crystal structure of a putative translation repressor from Vibrio cholerae
To be Published
|
|
3NEK
 
 | | Crystal structure of a nitrogen repressor-like protein MJ0159 from Methanococcus jannaschii | | Descriptor: | GLYCEROL, nitrogen repressor-like protein MJ0159 | | Authors: | Bonanno, J.B, Patskovsky, Y, Malashkevich, V, Ozyurt, S, Dickey, M, Wu, B, Maletic, M, Rodgers, L, Koss, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-09 | | Release date: | 2010-06-23 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural underpinnings of nitrogen regulation by the prototypical nitrogen-responsive transcriptional factor NrpR.
Structure, 18, 2010
|
|
2OCE
 
 | |
2I5G
 
 | | Crystal strcuture of amidohydrolase from Pseudomonas aeruginosa | | Descriptor: | amidohydrolase | | Authors: | Min, T, Sauder, J.M, Wasserman, S.R, Smith, D, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-24 | | Release date: | 2006-09-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of amidohydrolase from Pseudomonas aeruginosa
To be Published
|
|
2GUW
 
 | |
2HZT
 
 | | Crystal Structure of a putative HTH-type transcriptional regulator ytcD | | Descriptor: | Putative HTH-type transcriptional regulator ytcD | | Authors: | Madegowda, M, Eswaramoorthy, S, Desigan, K, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-09 | | Release date: | 2006-08-29 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a putative HTH-type transcription regulator ytcD
To be Published
|
|
2I5H
 
 | |
2KUC
 
 | | Solution Structure of a putative disulphide-isomerase from Bacteroides thetaiotaomicron | | Descriptor: | Putative disulphide-isomerase | | Authors: | Harris, R, Foti, R, Seidel, R.D, Bonanno, J.B, Freeman, J, Bain, K.T, Sauder, J.M, Burley, S.K, Girvin, M.E, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-17 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a putative disulphide-isomerase from Bacteroides thetaiotaomicron
To be Published
|
|
3N07
 
 | | Structure of putative 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase from Vibrio cholerae | | Descriptor: | 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase, MAGNESIUM ION | | Authors: | Liu, W, Ramagopal, U.A, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-13 | | Release date: | 2010-08-04 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural basis for the divergence of substrate specificity and biological function within HAD phosphatases in lipopolysaccharide and sialic acid biosynthesis.
Biochemistry, 52, 2013
|
|
3N7C
 
 | | Crystal structure of the RAN binding domain from the nuclear pore complex component NUP2 from Ashbya gossypii | | Descriptor: | ABR034Wp | | Authors: | Sampathkumar, P, Manglicmot, D, Gilmore, J, Bain, K, Gheyi, T, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-26 | | Release date: | 2010-06-16 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of the RAN binding domain from the nuclear pore complex component NUP2 from Ashbya gossypii
To be Published
|
|
3OVG
 
 | | The crystal structure of an amidohydrolase from Mycoplasma synoviae with Zn ion bound | | Descriptor: | PHOSPHATE ION, ZINC ION, amidohydrolase | | Authors: | Zhang, Z, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-09-16 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | The crystal structure of an amidohydrolase from Mycoplasma synoviae with Zn ion bound
To be Published
|
|
3OQB
 
 | | CRYSTAL STRUCTURE OF putative oxidoreductase from Bradyrhizobium japonicum USDA 110 | | Descriptor: | Oxidoreductase | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-09-02 | | Release date: | 2010-09-15 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CRYSTAL STRUCTURE OF putative oxidoreductase from Bradyrhizobium japonicum USDA 110
To be Published
|
|
3LQ7
 
 | | Crystal structure of glutathione s-transferase from agrobacterium tumefaciens str. c58 | | Descriptor: | Glutathione S-transferase | | Authors: | Patskovsky, Y, Toro, R, Gilmore, M, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-08 | | Release date: | 2010-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase from Agrobacterium Tumefaciens
To be Published
|
|
3P3D
 
 | | Crystal structure of the Nup53 RRM domain from Pichia guilliermondii | | Descriptor: | Nucleoporin 53 | | Authors: | Sampathkumar, P, Shawn, C, Bain, K, Gilmore, J, Gheyi, T, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-10-04 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the Nup53 RRM domain from Pichia guilliermondii
To be Published
|
|
3R03
 
 | |
3LMD
 
 | | Crystal structure of geranylgeranyl pyrophosphate synthase from corynebacterium glutamicum atcc 13032 | | Descriptor: | Geranylgeranyl pyrophosphate synthase | | Authors: | Patskovsky, Y, Ho, M, Toro, R, Rutter, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-01-29 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Geranylgeranyl Pyrophosphate Synthase from Corynebacterium Glutamicum
To be Published
|
|
3LPM
 
 | | Crystal structure of putative methyltransferase small domain protein from Listeria monocytogenes | | Descriptor: | Putative methyltransferase | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-05 | | Release date: | 2010-02-23 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of putative methyltransferase small domain protein from Listeria monocytogenes
To be Published
|
|
2QEE
 
 | | Crystal structure of putative amidohydrolase BH0493 from Bacillus halodurans C-125 | | Descriptor: | BH0493 protein, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Malashkevich, V.N, Toro, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-06-25 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of putative amidohydrolase BH0493 from Bacillus halodurans C-125.
To be Published
|
|
2QS8
 
 | | Crystal structure of a Xaa-Pro dipeptidase with bound methionine in the active site | | Descriptor: | MAGNESIUM ION, METHIONINE, Xaa-Pro Dipeptidase | | Authors: | Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-07-30 | | Release date: | 2007-08-21 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Functional annotation of two new carboxypeptidases from the amidohydrolase superfamily of enzymes.
Biochemistry, 48, 2009
|
|
2NQL
 
 | | Crystal structure of a member of the enolase superfamily from Agrobacterium tumefaciens | | Descriptor: | GLYCEROL, Isomerase/lactonizing enzyme, SODIUM ION, ... | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Sauder, J.M, Dickey, M, Adams, J.M, Ozyurt, S, Wasserman, S.R, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-31 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Enolase from Agrobacterium Tumefaciens C58
To be Published
|
|
3LGX
 
 | |
3LNV
 
 | | The crystal structure of fatty acyl-adenylate ligase from L. pneumophila in complex with acyl adenylate and pyrophosphate | | Descriptor: | 5'-O-[(S)-(dodecanoyloxy)(hydroxy)phosphoryl]adenosine, PYROPHOSPHATE 2-, Saframycin Mx1 synthetase B | | Authors: | Zhang, Z, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-03 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Studies of Fatty Acyl Adenylate Ligases from E. coli and L. pneumophila.
J.Mol.Biol., 406, 2011
|
|
3LOP
 
 | | Crystal structure of substrate-binding periplasmic protein (Pbp) from Ralstonia solanacearum | | Descriptor: | 1,2-ETHANEDIOL, LEUCINE, MAGNESIUM ION, ... | | Authors: | Palani, K, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-04 | | Release date: | 2010-02-23 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of substrate-binding periplasmic protein (Pbp) from Ralstonia solanacearum
To be Published
|
|
3LSZ
 
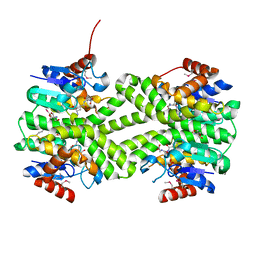 | | Crystal structure of glutathione s-transferase from Rhodobacter sphaeroides | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLUTATHIONE, GLYCEROL, ... | | Authors: | Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-14 | | Release date: | 2010-03-23 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of glutathione s-transferase from Rhodobacter sphaeroides
To be Published
|
|
