6E5T
 
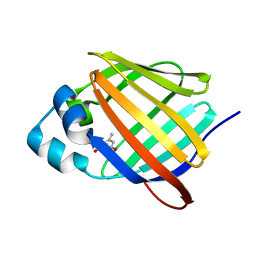 | | Crystal structure of human cellular retinol binding protein 1 in complex with abnormal-cannabidiorcin (Abn-CBDO) | | Descriptor: | (1'R,2'R)-5',6-dimethyl-2'-(prop-1-en-2-yl)-1',2',3',4'-tetrahydro[1,1'-biphenyl]-2,4-diol, Retinol-binding protein 1 | | Authors: | Silvaroli, J.A, Horwitz, S, Banerjee, S, Kiser, P.D, Golczak, M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Abnormal Cannabidiol Modulates Vitamin A Metabolism by Acting as a Competitive Inhibitor of CRBP1.
Acs Chem.Biol., 14, 2019
|
|
5W58
 
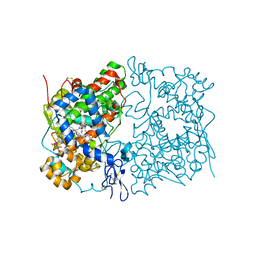 | | Crystal Complex of Cyclooxygenase-2: (S)-ARN-2508 (a dual COX and FAAH inhibitor) | | Descriptor: | (2S)-2-{2-fluoro-3'-[(hexylcarbamoyl)oxy][1,1'-biphenyl]-4-yl}propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, S, Goodman, M.C, Banerjee, S, Piomelli, D, Marnett, L.J. | | Deposit date: | 2017-06-14 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.267 Å) | | Cite: | Dual cyclooxygenase-fatty acid amide hydrolase inhibitor exploits novel binding interactions in the cyclooxygenase active site.
J. Biol. Chem., 293, 2018
|
|
6X6O
 
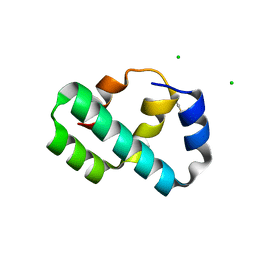 | | Crystal structure of T4 protein Spackle as determined by native SAD phasing | | Descriptor: | CHLORIDE ION, Protein spackle | | Authors: | Shi, K, Kurniawan, F, Banerjee, S, Moeller, N.H, Aihara, H. | | Deposit date: | 2020-05-28 | | Release date: | 2020-09-16 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structure of bacteriophage T4 Spackle as determined by native SAD phasing.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6RA3
 
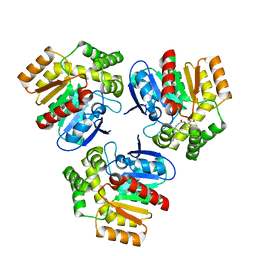 | | Structural basis for recognition and ring-cleavage of the Pseudomonas quinolone signal (PQS) by AqDC in complex with its product | | Descriptor: | 2-(octanoylamino)benzoic acid, Putative dioxygenase (1H-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase) | | Authors: | Wullich, S, Kobus, S, Smits, S.H, Fetzner, S. | | Deposit date: | 2019-04-05 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for recognition and ring-cleavage of the Pseudomonas quinolone signal (PQS) by AqdC, a mycobacterial dioxygenase of the alpha / beta-hydrolase fold family.
J.Struct.Biol., 207, 2019
|
|
1ECV
 
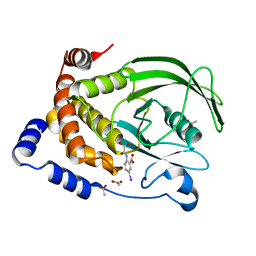 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH 5-IODO-2-(OXALYL-AMINO)-BENZOIC ACID | | Descriptor: | 5-IODO-2-(OXALYL-AMINO)-BENZOIC ACID, ACETATE ION, PROTEIN-TYROSINE PHOSPHATASE 1B | | Authors: | Andersen, H.S, Iversen, L.F, Branner, S, Rasmussen, H.B, Moller, N.P.H. | | Deposit date: | 2000-01-26 | | Release date: | 2000-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 2-(oxalylamino)-benzoic acid is a general, competitive inhibitor of protein-tyrosine phosphatases.
J.Biol.Chem., 275, 2000
|
|
6RA2
 
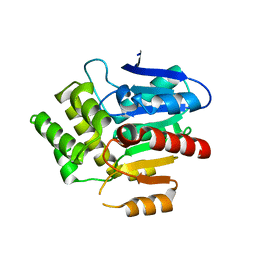 | | Structural basis for recognition and ring-cleavage of the Pseudomonas quinolone signal (PQS) by AqDC | | Descriptor: | Putative dioxygenase (1H-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase) | | Authors: | Wullich, S, Kobus, S, Smits, S.H, Fetzner, S. | | Deposit date: | 2019-04-05 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for recognition and ring-cleavage of the Pseudomonas quinolone signal (PQS) by AqdC, a mycobacterial dioxygenase of the alpha / beta-hydrolase fold family.
J.Struct.Biol., 207, 2019
|
|
5A6Y
 
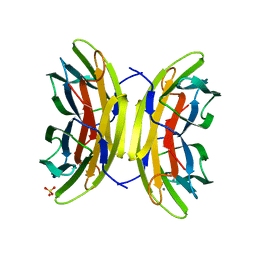 | | Structure of the LecB lectin from Pseudomonas aeruginosa strain PA14 in complex with mannose-alpha1,3mannoside | | Descriptor: | CALCIUM ION, FUCOSE-BINDING LECTIN PA-IIL, GLYCEROL, ... | | Authors: | Sommer, R, Wagner, S, Varrot, A, Khaledi, A, Haussler, S, Imberty, A, Titz, A. | | Deposit date: | 2015-07-02 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The virulence factor LecB varies in clinical isolates: consequences for ligand binding and drug discovery.
Chem Sci, 7, 2016
|
|
5A70
 
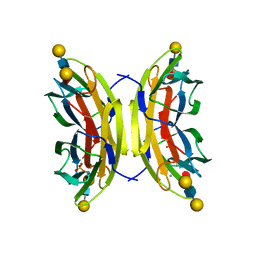 | | Structure of the LecB lectin from Pseudomonas aeruginosa strain PA14 in complex with lewis x tetrasaccharide | | Descriptor: | CALCIUM ION, LECB, SULFATE ION, ... | | Authors: | Sommer, R, Wagner, S, Varrot, A, Khaledi, A, Haussler, S, Imberty, A, Titz, A. | | Deposit date: | 2015-07-02 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Induction of rare conformation of oligosaccharide by binding to calcium-dependent bacterial lectin: X-ray crystallography and modelling study.
Eur.J.Med.Chem., 177, 2019
|
|
5I7V
 
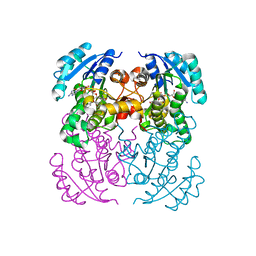 | | Crystal structure of B. pseudomallei FabI in complex with NAD and PT02 | | Descriptor: | 2-phenoxy-5-propyl-phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Hirschbeck, M.W, Eltschkner, S, Tonge, P.J, Kisker, C. | | Deposit date: | 2016-02-18 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Rationalizing the Binding Kinetics for the Inhibition of the Burkholderia pseudomallei FabI1 Enoyl-ACP Reductase.
Biochemistry, 56, 2017
|
|
4GC6
 
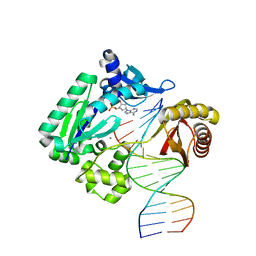 | | Crystal structure of Dpo4 in complex with N-MC-dAMP opposite dT | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*CP*C)-3'), DNA (5'-D(*TP*CP*AP*TP*GP*GP*AP*AP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3'), ... | | Authors: | Eoff, R.L, Ketkar, A, Banerjee, S, Zafar, M.K. | | Deposit date: | 2012-07-29 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.895 Å) | | Cite: | Differential furanose selection in the active sites of archaeal DNA polymerases probed by fixed-conformation nucleotide analogues.
Biochemistry, 51, 2012
|
|
5I9L
 
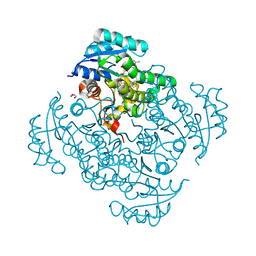 | | Crystal structure of B. pseudomallei FabI in complex with NAD and PT404 | | Descriptor: | 2-(2-chloro-4-nitrophenoxy)-5-ethyl-4-fluorophenol, Enoyl-[acyl-carrier-protein] reductase [NADH], GLYCEROL, ... | | Authors: | Hirschbeck, M.W, Eltschkner, S, Tonge, P.J, Kisker, C. | | Deposit date: | 2016-02-20 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rationalizing the Binding Kinetics for the Inhibition of the Burkholderia pseudomallei FabI1 Enoyl-ACP Reductase.
Biochemistry, 56, 2017
|
|
5I8Z
 
 | | Crystal structure of B. pseudomallei FabI in complex with NAD and PT12 | | Descriptor: | 5-HEXYL-2-(4-NITROPHENOXY)PHENOL, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Hirschbeck, M.W, Eltschkner, S, Tonge, P.J, Kisker, C. | | Deposit date: | 2016-02-19 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.623 Å) | | Cite: | Rationalizing the Binding Kinetics for the Inhibition of the Burkholderia pseudomallei FabI1 Enoyl-ACP Reductase.
Biochemistry, 56, 2017
|
|
5A6X
 
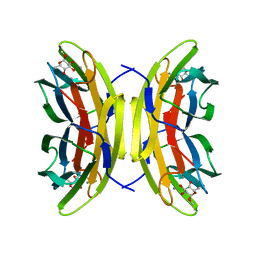 | | Structure of the LecB lectin from Pseudomonas aeruginosa strain PA14 in complex with alpha-methyl-fucoside | | Descriptor: | CALCIUM ION, FUCOSE-BINDING LECTIN PA-IIL, methyl alpha-L-fucopyranoside | | Authors: | Sommer, R, Wagner, S, Varrot, A, Khaledi, A, Haussler, S, Imberty, A, Titz, A. | | Deposit date: | 2015-07-02 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The virulence factor LecB varies in clinical isolates: consequences for ligand binding and drug discovery.
Chem Sci, 7, 2016
|
|
5IFL
 
 | | Crystal structure of B. pseudomallei FabI in complex with NAD and triclosan | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Hirschbeck, M.W, Eltschkner, S, Tonge, P.J, Kisker, C. | | Deposit date: | 2016-02-26 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Rationalizing the Binding Kinetics for the Inhibition of the Burkholderia pseudomallei FabI1 Enoyl-ACP Reductase.
Biochemistry, 56, 2017
|
|
5I8W
 
 | | Crystal structure of B. pseudomallei FabI in complex with NAD and PT401 | | Descriptor: | 4-fluoro-5-hexyl-2-(2-methylphenoxy)phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Hirschbeck, M.W, Eltschkner, S, Tonge, P.J, Kisker, C. | | Deposit date: | 2016-02-19 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | Rationalizing the Binding Kinetics for the Inhibition of the Burkholderia pseudomallei FabI1 Enoyl-ACP Reductase.
Biochemistry, 56, 2017
|
|
5I7S
 
 | | Crystal structure of B. pseudomallei FabI in complex with NAD and PT01 | | Descriptor: | 5-ETHYL-2-PHENOXYPHENOL, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Hirschbeck, M.W, Eltschkner, S, Tonge, P.J, Kisker, C. | | Deposit date: | 2016-02-18 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.595 Å) | | Cite: | Rationalizing the Binding Kinetics for the Inhibition of the Burkholderia pseudomallei FabI1 Enoyl-ACP Reductase.
Biochemistry, 56, 2017
|
|
1N1M
 
 | | Human Dipeptidyl Peptidase IV/CD26 in complex with an inhibitor | | Descriptor: | 2-AMINO-3-METHYL-1-PYRROLIDIN-1-YL-BUTAN-1-ONE, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rasmussen, H.B, Branner, S, Wiberg, F.C, Wagtmann, N.R. | | Deposit date: | 2002-10-18 | | Release date: | 2002-12-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human dipeptidyl peptidase IV/CD26 in complex with a substrate analogue
Nat.Struct.Biol., 10, 2003
|
|
4UYR
 
 | | X-ray structure of the N-terminal domain of the flocculin Flo11 from Saccharomyces cerevisiae | | Descriptor: | FLOCCULATION PROTEIN FLO11, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Veelders, M, Kraushaar, T, Brueckner, S, Rhinow, D, Moesch, H.U, Essen, L.O. | | Deposit date: | 2014-09-03 | | Release date: | 2015-08-12 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Interactions by the Fungal Flo11 Adhesin Depend on a Fibronectin Type III-Like Adhesin Domain Girdled by Aromatic Bands.
Structure, 23, 2015
|
|
4UYS
 
 | | X-ray structure of the N-terminal domain of the flocculin Flo11 from Saccharomyces cerevisiae | | Descriptor: | FLOCCULATION PROTEIN FLO11, MAGNESIUM ION, SODIUM ION | | Authors: | Kraushaar, T, Veelders, M, Brueckner, S, Rhinow, D, Moesch, H.U, Essen, L.O. | | Deposit date: | 2014-09-03 | | Release date: | 2015-08-12 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Interactions by the Fungal Flo11 Adhesin Depend on a Fibronectin Type III-Like Adhesin Domain Girdled by Aromatic Bands.
Structure, 23, 2015
|
|
7ZE9
 
 | | Structure of an AA16 LPMO-like protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, COPPER (II) ION, ... | | Authors: | Huang, Z, Banerjee, S, Muderspach, S.J, Sun, P, van Berkel, W.J.H, Kabel, M.A, Lo Leggio, L. | | Deposit date: | 2022-03-30 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.646 Å) | | Cite: | AA16 Oxidoreductases Boost Cellulose-Active AA9 Lytic Polysaccharide Monooxygenases from Myceliophthora thermophila.
Acs Catalysis, 13, 2023
|
|
4ZSI
 
 | | Crystal structure of the effector-binding domain of DasR (DasR-EBD) in complex with glucosamine-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, 2-amino-2-deoxy-6-O-phosphono-beta-D-glucopyranose, ... | | Authors: | Fillenberg, S.B, Koerner, S, Muller, Y.A. | | Deposit date: | 2015-05-13 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | Crystal Structures of the Global Regulator DasR from Streptomyces coelicolor: Implications for the Allosteric Regulation of GntR/HutC Repressors.
Plos One, 11, 2016
|
|
5A3L
 
 | | Structure of Cea1A in complex with N-Acetylglucosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Kock, M, Brueckner, S, Wozniak, N, Veelders, M, Schlereth, J, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2015-06-02 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | High-Affinity Recognition of Non-Reducing Chitinous Ends by the Yeast Adhesin Cea1
To be Published
|
|
5ILO
 
 | | Crystal structure of photoreceptor dehydrogenase from Drosophila melanogaster | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Photoreceptor dehydrogenase, isoform C | | Authors: | Hofmann, L, Tsybovsky, Y, Banerjee, S. | | Deposit date: | 2016-03-04 | | Release date: | 2016-11-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural Insights into the Drosophila melanogaster Retinol Dehydrogenase, a Member of the Short-Chain Dehydrogenase/Reductase Family.
Biochemistry, 55, 2016
|
|
5IFO
 
 | | X-ray structure of HSA-Myr-KP1019 | | Descriptor: | MYRISTIC ACID, RUTHENIUM ION, Serum albumin | | Authors: | Bijelic, A, Theiner, S, Keppler, B.K, Rompel, A. | | Deposit date: | 2016-02-26 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray Structure Analysis of Indazolium trans-[Tetrachlorobis(1H-indazole)ruthenate(III)] (KP1019) Bound to Human Serum Albumin Reveals Two Ruthenium Binding Sites and Provides Insights into the Drug Binding Mechanism.
J.Med.Chem., 59, 2016
|
|
5ILG
 
 | | Crystal structure of photoreceptor dehydrogenase from Drosophila melanogaster | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Hofmann, L, Tsybovsky, Y, Banerjee, S. | | Deposit date: | 2016-03-04 | | Release date: | 2016-11-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights into the Drosophila melanogaster Retinol Dehydrogenase, a Member of the Short-Chain Dehydrogenase/Reductase Family.
Biochemistry, 55, 2016
|
|
