3BE3
 
 | |
3K2G
 
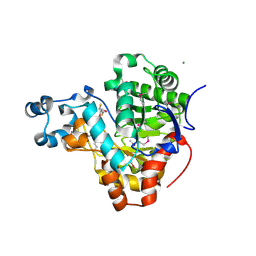 | | Crystal structure of a Resiniferatoxin-binding protein from Rhodobacter sphaeroides | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, MAGNESIUM ION, Resiniferatoxin-binding, ... | | Authors: | Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-30 | | Release date: | 2009-10-13 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a Resiniferatoxin-binding protein from Rhodobacter sphaeroides
To be Published
|
|
3BWI
 
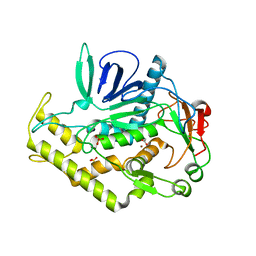 | | Crystal structure of the catalytic domain of botulinum neurotoxin serotype A with an acetate ion bound at the active site | | Descriptor: | ACETATE ION, Botulinum neurotoxin A light chain, SULFATE ION, ... | | Authors: | Kumaran, D, Rawat, R, Swaminathan, S. | | Deposit date: | 2008-01-09 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure- and Substrate-based Inhibitor Design for Clostridium botulinum Neurotoxin Serotype A
J.Biol.Chem., 283, 2008
|
|
3C88
 
 | |
2PZZ
 
 | |
3BT3
 
 | |
3KXW
 
 | |
2A9F
 
 | |
3GPK
 
 | |
3EHE
 
 | |
2QZJ
 
 | |
3E9M
 
 | |
3HDG
 
 | |
3HDP
 
 | |
3IH0
 
 | | Crystal structure of an uncharacterized sugar kinase PH1459 from Pyrococcus horikoshii in complex with AMP-PNP | | Descriptor: | GLYCEROL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Uncharacterized sugar kinase PH1459 | | Authors: | Kumar, G, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-29 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an uncharacterized sugar kinase PH1459 from Pyrococcus horikoshii in complex with AMP-PNP
To be Published
|
|
3C8B
 
 | |
3C89
 
 | |
3D5L
 
 | |
3DLI
 
 | |
3EXQ
 
 | |
3HP0
 
 | |
3H49
 
 | |
3DDA
 
 | |
3DDB
 
 | |
3FIJ
 
 | |
