3OC4
 
 | | Crystal Structure of a pyridine nucleotide-disulfide family oxidoreductase from the Enterococcus faecalis V583 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Oxidoreductase, pyridine nucleotide-disulfide family, ... | | Authors: | Kumaran, D, Baumann, K, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-08-09 | | Release date: | 2010-10-06 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of a pyridine nucleotide-disulfide family oxidoreductase from the Enterococcus faecalis V583
To be Published
|
|
3NQB
 
 | |
1YBE
 
 | |
2NWU
 
 | |
3D3X
 
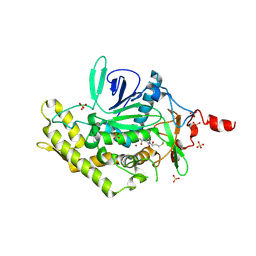 | |
2RBB
 
 | |
2RK0
 
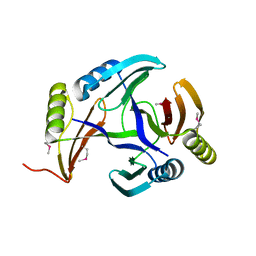 | |
2F4N
 
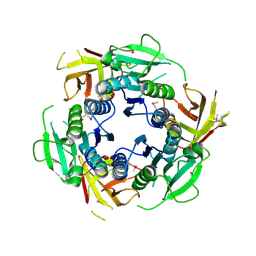 | |
3PZL
 
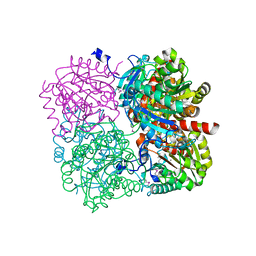 | |
2RGY
 
 | |
3QDK
 
 | |
3N87
 
 | | Crystal structure of 3-dehydroquinate dehydratase from Mycobacterium tuberculosis in complex with inhibitor 3 | | Descriptor: | (1R,4R,5R)-1,4,5-trihydroxy-3-[3-(phenylcarbonyl)phenyl]cyclohex-2-ene-1-carboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Dias, M.V.B, Snee, W.C, Bromfield, K.M, Payne, R, Palaninathan, S.K, Ciulli, A, Howard, N.I, Abell, C, Sacchettini, J.C, Blundell, T.L. | | Deposit date: | 2010-05-27 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural investigation of inhibitor designs targeting 3-dehydroquinate dehydratase from the shikimate pathway of Mycobacterium tuberculosis.
Biochem.J., 436, 2011
|
|
3DCN
 
 | | Glomerella cingulata apo cutinase | | Descriptor: | Cutinase | | Authors: | Nyon, M.P, Rice, D.W, Berrisford, J.M, Hounslow, A.M, Moir, A.J.G, Huang, H, Nathan, S, Mahadi, N.M, Farah Diba, A.B, Craven, C.J. | | Deposit date: | 2008-06-04 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalysis by Glomerella cingulata Cutinase Requires Conformational Cycling between the Active and Inactive States of Its Catalytic Triad
J.Mol.Biol., 385, 2009
|
|
3DEA
 
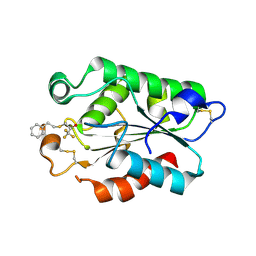 | | Glomerella cingulata PETFP-cutinase complex | | Descriptor: | 1,1,1-trifluoro-3-[(2-phenylethyl)sulfanyl]propan-2-one, Cutinase | | Authors: | Nyon, M.P, Rice, D.W, Berrisford, J.M, Hounslow, A.M, Moir, A.J.G, Huang, H, Nathan, S, Mahadi, N.M, Farah Diba, A.B, Craven, C.J. | | Deposit date: | 2008-06-09 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Catalysis by Glomerella cingulata Cutinase Requires Conformational Cycling between the Active and Inactive States of Its Catalytic Triad
J.Mol.Biol., 385, 2009
|
|
3QW6
 
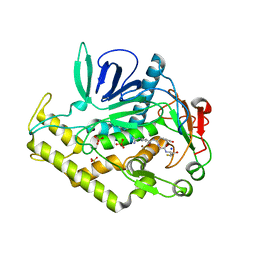 | | Crystal structure of the protease domain of Botulinum Neurotoxin Serotype A with a peptide inhibitor RYGC | | Descriptor: | Botulinum neurotoxin type A, SODIUM ION, SULFATE ION, ... | | Authors: | Kumaran, D, Swaminathan, S. | | Deposit date: | 2011-02-26 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Peptide inhibitors of botulinum neurotoxin serotype A: design, inhibition, cocrystal structures, structure-activity relationship and pharmacophore modeling.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1YBD
 
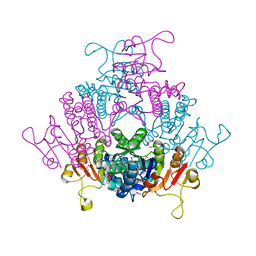 | |
3QW7
 
 | | Crystal structure of the protease domain of Botulinum Neurotoxin Serotype A with a peptide inhibitor RRFC | | Descriptor: | Botulinum neurotoxin type A, SODIUM ION, SULFATE ION, ... | | Authors: | Kumaran, D, Swaminathan, S. | | Deposit date: | 2011-02-27 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Peptide inhibitors of botulinum neurotoxin serotype A: design, inhibition, cocrystal structures, structure-activity relationship and pharmacophore modeling.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3NWM
 
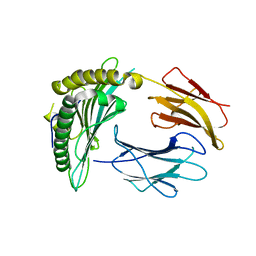 | | Crystal structure of a single chain construct composed of MHC class I H-2Kd, beta-2microglobulin and a peptide which is an autoantigen for type 1 diabetes | | Descriptor: | Peptide/beta-2microglobulin/MHC class I H-2Kd chimeric protein | | Authors: | Ramagopal, U.A, Samanta, D, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2010-07-09 | | Release date: | 2011-07-06 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional characterization of a single-chain peptide-MHC molecule that modulates both naive and activated CD8+ T cells.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3NO1
 
 | |
2PKD
 
 | | Crystal structure of CD84: Insite into SLAM family function | | Descriptor: | CHLORIDE ION, SLAM family member 5 | | Authors: | Yan, Q, Malashkevich, V.N, Fedorov, A, Cao, E, Lary, J.W, Cole, J.L, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2007-04-17 | | Release date: | 2007-06-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.043 Å) | | Cite: | Structure of CD84 provides insight into SLAM family function.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2GOK
 
 | | Crystal structure of the imidazolonepropionase from Agrobacterium tumefaciens at 1.87 A resolution | | Descriptor: | CHLORIDE ION, FE (III) ION, GLYCEROL, ... | | Authors: | Tyagi, R, Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-04-13 | | Release date: | 2006-04-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | X-ray structure of imidazolonepropionase from Agrobacterium tumefaciens at 1.87 A resolution.
Proteins, 69, 2007
|
|
3QW5
 
 | | Crystal structure of the protease domain of Botulinum Neurotoxin Serotype A with a peptide inhibitor RRGF | | Descriptor: | Botulinum neurotoxin type A, SULFATE ION, ZINC ION, ... | | Authors: | Kumaran, D, Swaminathan, S. | | Deposit date: | 2011-02-26 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Peptide inhibitors of botulinum neurotoxin serotype A: design, inhibition, cocrystal structures, structure-activity relationship and pharmacophore modeling.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3QW8
 
 | | Crystal structure of the protease domain of Botulinum Neurotoxin Serotype A with a peptide inhibitor CRGC | | Descriptor: | 1,2-ETHANEDIOL, Botulinum neurotoxin type A, SODIUM ION, ... | | Authors: | Kumaran, D, Swaminathan, S. | | Deposit date: | 2011-02-27 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Peptide inhibitors of botulinum neurotoxin serotype A: design, inhibition, cocrystal structures, structure-activity relationship and pharmacophore modeling.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3R0N
 
 | | Crystal Structure of the Immunoglobulin variable domain of Nectin-2 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Poliovirus receptor-related protein 2 | | Authors: | Ramagopal, U.A, Samanta, D, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2011-03-08 | | Release date: | 2011-04-27 | | Last modified: | 2012-11-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of Nectin-2 reveals determinants of homophilic and heterophilic interactions that control cell-cell adhesion.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3RP1
 
 | | Crystal structure of Human LAIR-1 in C2 space group | | Descriptor: | Leukocyte-associated immunoglobulin-like receptor 1 | | Authors: | Sampathkumar, P, Ramagopal, U.A, Yan, Q, Toro, R, Nathenson, S, Bonanno, J, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-04-26 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Human LAIR-1 in C2 space group
To be Published
|
|
