1CQV
 
 | |
8EKO
 
 | |
4S3I
 
 | |
2A9F
 
 | |
3DH0
 
 | |
4QXD
 
 | | Crystal structure of Inositol Polyphosphate 1-Phosphatase from Entamoeba histolytica | | Descriptor: | 3'(2'),5'-bisphosphate nucleotidase, putative, MAGNESIUM ION, ... | | Authors: | Tarique, K.F, Abdul Rehman, S.A, Betzel, C, Gourinath, S. | | Deposit date: | 2014-07-19 | | Release date: | 2014-08-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure-based identification of inositol polyphosphate 1-phosphatase from Entamoeba histolytica
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3GPK
 
 | |
3EAF
 
 | | Crystal structure of ABC transporter, substrate binding protein Aeropyrum pernix | | Descriptor: | ABC transporter, substrate binding protein, GLYCEROL, ... | | Authors: | Zhang, Z, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-25 | | Release date: | 2008-09-09 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of ABC transporter, substrate binding protein Aeropyrum pernix
To be Published
|
|
3EWM
 
 | |
3H49
 
 | |
3HDP
 
 | |
3EHE
 
 | |
3E9M
 
 | |
3EXQ
 
 | |
3FIJ
 
 | |
4L1E
 
 | | Crystal structure of C-Phycocyanin from Leptolyngbya sp. N62DM | | Descriptor: | BILIVERDINE IX ALPHA, PHYCOCYANOBILIN, Phycocyanin alpha chain, ... | | Authors: | Singh, N.K, Raj, I, Gourinath, S, Madamwar, D. | | Deposit date: | 2013-06-03 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal Structure and Interaction of Phycocyanin with beta-Secretase: A Putative Therapy for Alzheimer's Disease.
CNS Neurol Disord Drug Targets, 13, 2014
|
|
3DZB
 
 | |
3DLI
 
 | |
1CCD
 
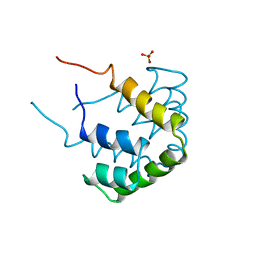 | | REFINED STRUCTURE OF RAT CLARA CELL 17 KDA PROTEIN AT 3.0 ANGSTROMS RESOLUTION | | Descriptor: | CLARA CELL 17 kD PROTEIN, SULFATE ION | | Authors: | Umland, T.C, Swaminathan, S, Furey, W, Singh, G, Pletcher, J, Sax, M. | | Deposit date: | 1991-09-17 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Refined structure of rat Clara cell 17 kDa protein at 3.0 A resolution.
J.Mol.Biol., 224, 1992
|
|
3DDB
 
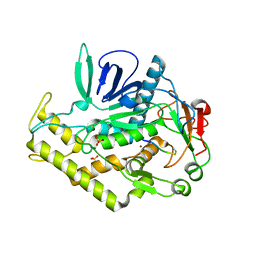 | |
1Z2L
 
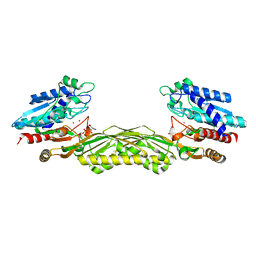 | | Crystal structure of Allantoate-amidohydrolase from E.coli K12 in complex with substrate Allantoate | | Descriptor: | ALLANTOATE ION, Allantoate amidohydrolase, SULFATE ION, ... | | Authors: | Agarwal, R, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-03-08 | | Release date: | 2005-03-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural analysis of a ternary complex of allantoate amidohydrolase from Escherichia coli reveals its mechanics.
J.Mol.Biol., 368, 2007
|
|
5DA3
 
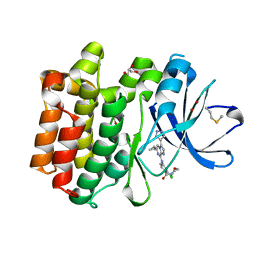 | | Crystal structure of PTK6 Kinase domain with inhibitor | | Descriptor: | (2-chloro-4-{[6-cyclopropyl-3-(1H-pyrazol-4-yl)imidazo[1,2-a]pyrazin-8-yl]amino}phenyl)(morpholin-4-yl)methanone, GLYCEROL, Protein-tyrosine kinase 6 | | Authors: | Thakur, M.K, Birudukota, S, Swaminathan, S, Battula, S.K, Vadivelu, S, Tyagi, R, Gosu, R. | | Deposit date: | 2015-08-19 | | Release date: | 2016-08-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Co-crystal structures of PTK6: With Dasatinib at 2.24 angstrom , with novel imidazo[1,2-a]pyrazin-8-amine derivative inhibitor at 1.70 angstrom resolution
Biochem. Biophys. Res. Commun., 482, 2017
|
|
1ZCC
 
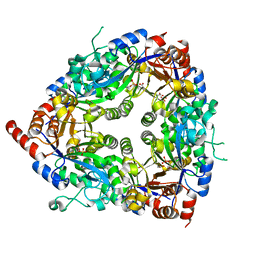 | | Crystal structure of glycerophosphodiester phosphodiesterase from Agrobacterium tumefaciens str.C58 | | Descriptor: | ACETATE ION, SULFATE ION, glycerophosphodiester phosphodiesterase | | Authors: | Krishnamurthy, N.R, Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-04-11 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of glycerophosphodiester phosphodiesterase from Agrobacterium tumefaciens by SAD with a large asymmetric unit.
Proteins, 65, 2006
|
|
5G4Q
 
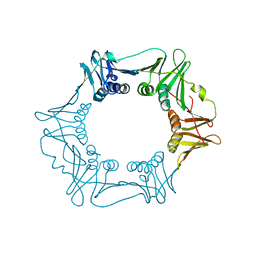 | | H.pylori Beta clamp in complex with 5-chloroisatin | | Descriptor: | 5-chloro-1H-indole-2,3-dione, DNA POLYMERASE III SUBUNIT BETA | | Authors: | Pandey, P, Gourinath, S. | | Deposit date: | 2016-05-16 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Screening of E. coli beta-clamp Inhibitors Revealed that Few Inhibit Helicobacter pylori More Effectively: Structural and Functional Characterization.
Antibiotics (Basel), 7, 2018
|
|
5G48
 
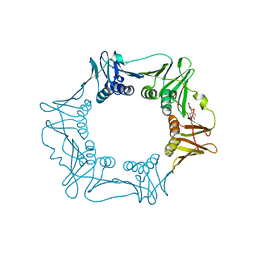 | | H.pylori Beta clamp in complex with Diflunisal | | Descriptor: | 5-(2,4-DIFLUOROPHENYL)-2-HYDROXY-BENZOIC ACID, DNA POLYMERASE III SUBUNIT BETA | | Authors: | Pandey, P, Gourinath, S. | | Deposit date: | 2016-05-06 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Targeting the beta-clamp in Helicobacter pylori with FDA-approved drugs reveals micromolar inhibition by diflunisal.
FEBS Lett., 591, 2017
|
|
