5EY7
 
 | |
5F11
 
 | |
5EYN
 
 | | Crystal structure of Fructokinase from Vibrio cholerae O395 in fructose, ADP, Beryllium trifluoride and calcium ion bound form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, CALCIUM ION, ... | | Authors: | Paul, R, Nath, S, Sen, U. | | Deposit date: | 2015-11-25 | | Release date: | 2016-11-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.289 Å) | | Cite: | Crystal structure of Fructokinase from Vibrio cholerae O395 in apo form
To Be Published
|
|
5F0Z
 
 | |
5Z3M
 
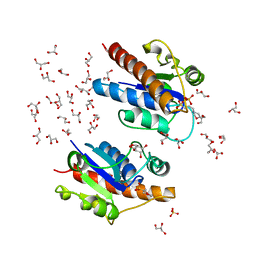 | | Crystal structure of Low Molecular Weight Phosphotyrosine phosphatase (VcLMWPTP-2) from Vibrio choleraeO395 | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, GLYCEROL, Phosphotyrosine protein phosphatase, ... | | Authors: | Chatterjee, S, Nath, S, Sen, U. | | Deposit date: | 2018-01-08 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Vibrio cholerae LMWPTP-2 display unique surface charge and grooves around the active site: Indicative of distinctive substrate specificity and scope to design specific inhibitor.
Biochim Biophys Acta Proteins Proteom, 1867, 2019
|
|
4DVD
 
 | |
5B74
 
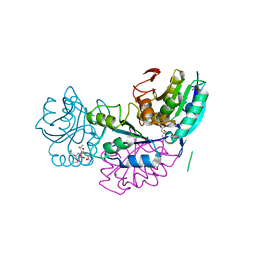 | | Crystal structure of conjoined Pyrococcus furiosus L-asparaginase with peptide | | Descriptor: | CHLORIDE ION, CITRATE ANION, L-asparaginase, ... | | Authors: | Sharma, P, Tomar, R, Nath, S.K, Kundu, B, Ashish, F. | | Deposit date: | 2016-06-05 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.039 Å) | | Cite: | Heat induces end to end repetitive association in P. furiosus L-asparaginase which enables its thermophilic property.
Sci Rep, 10, 2020
|
|
5B5U
 
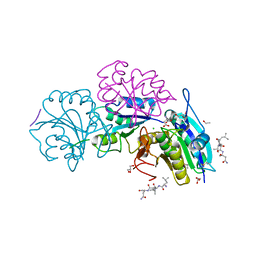 | | Crystal structure of truncated Pyrococcus furiosus L-asparaginase with peptide | | Descriptor: | ASPARTIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sharma, P, Tomar, R, Nath, S.K, Kundu, B, Ashish, F. | | Deposit date: | 2016-05-23 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Heat induces end to end repetitive association in P. furiosus L-asparaginase which enables its thermophilic property.
Sci Rep, 10, 2020
|
|
3HKF
 
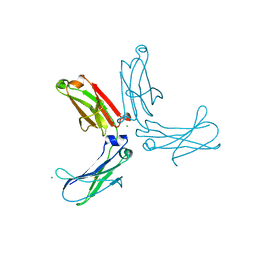 | | Murine unglycosylated IgG Fc fragment | | Descriptor: | CHLORIDE ION, Igh protein, MAGNESIUM ION | | Authors: | Feige, M.J, Nath, S, Catharino, S.R, Weinfurtner, D, Steinbacher, S, Buchner, J. | | Deposit date: | 2009-05-23 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the murine unglycosylated IgG1 Fc fragment
J.Mol.Biol., 391, 2009
|
|
5D7V
 
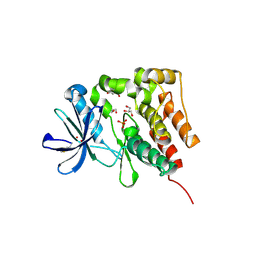 | | Crystal structure of PTK6 kinase domain | | Descriptor: | GLYCEROL, PHOSPHATE ION, Protein-tyrosine kinase 6 | | Authors: | Thakur, M.K, Birudukota, S, Swaminathan, S, Tyagi, R, Gosu, R. | | Deposit date: | 2015-08-14 | | Release date: | 2016-08-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of the kinase domain of human protein tyrosine kinase 6 (PTK6) at 2.33 angstrom resolution
Biochem.Biophys.Res.Commun., 478, 2016
|
|
8I11
 
 | |
8IL9
 
 | |
4IM9
 
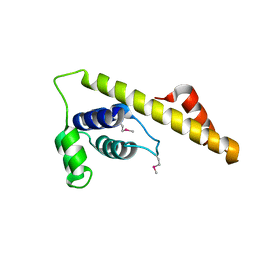 | |
4IL5
 
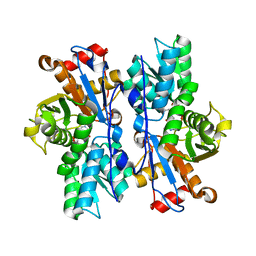 | |
3EB2
 
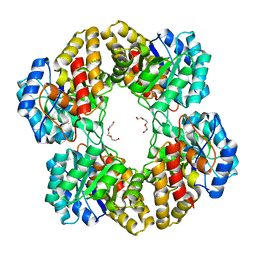 | |
2ISN
 
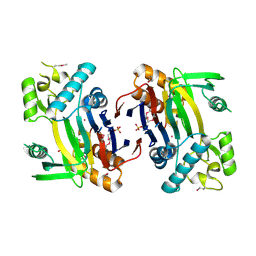 | | Crystal structure of a phosphatase from a pathogenic strain Toxoplasma gondii | | Descriptor: | NYSGXRC-8828z, phosphatase, PRASEODYMIUM ION, ... | | Authors: | Agarwal, R, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-18 | | Release date: | 2006-10-31 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
4S1I
 
 | | Pyridoxal Kinase of Entamoeba histolytica with PLP | | Descriptor: | MAGNESIUM ION, PYRIDOXAL-5'-PHOSPHATE, Pyridoxal kinase | | Authors: | Tarique, K.F, Devi, S, Abdul Rehman, S.A, Gourinath, S. | | Deposit date: | 2015-01-14 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Characterization and functional insights into the Entamoeba histolytica pyridoxal kinase, an enzyme essential for its survival.
J.Struct.Biol., 212, 2020
|
|
4S1M
 
 | | Crystal Structure of Pyridoxal Kinase from Entamoeba histolytica | | Descriptor: | MAGNESIUM ION, Pyridoxal kinase | | Authors: | Tarique, K.F, Devi, S, Abdul Rehman, S.A, Gourinath, S. | | Deposit date: | 2015-01-14 | | Release date: | 2015-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Characterization and functional insights into the Entamoeba histolytica pyridoxal kinase, an enzyme essential for its survival.
J.Struct.Biol., 212, 2020
|
|
3E9M
 
 | |
3E3V
 
 | |
2I6E
 
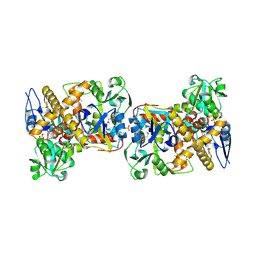 | | Crystal structure of protein DR0370 from Deinococcus radiodurans, Pfam DUF178 | | Descriptor: | Hypothetical protein, SULFATE ION | | Authors: | Tyagi, R, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-28 | | Release date: | 2006-09-05 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structures of two proteins belonging to Pfam DUF178 revealed unexpected structural similarity to the DUF191 Pfam family.
Bmc Struct.Biol., 7, 2007
|
|
2I76
 
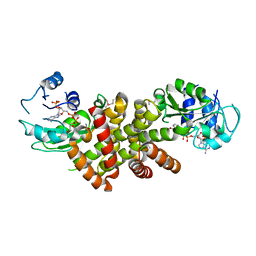 | | Crystal structure of protein TM1727 from Thermotoga maritima | | Descriptor: | Hypothetical protein, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Madegowda, M, Eswaramoorthy, S, Seetharaman, J, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-30 | | Release date: | 2006-10-03 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of hypothetical protein TM1727 from Thermatoga maritima
TO BE PUBLISHED
|
|
2I3O
 
 | |
3EUW
 
 | |
4RKI
 
 | |
