4RZ8
 
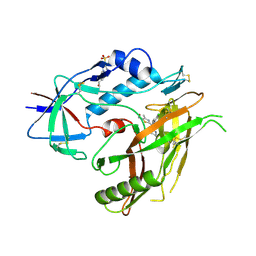 | | Crystal structure of HIV-1 gp120 core in complex with NBD-11021, a small molecule CD4-antagonist | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-(4-chlorophenyl)-N-{(S)-[5-(hydroxymethyl)-4-methyl-1,3-thiazol-2-yl][(2R)-piperidin-2-yl]methyl}-1H-pyrrole-2-carboxamide, ... | | Authors: | Kwon, Y.D, Debnath, A.K, Kwong, P.D. | | Deposit date: | 2014-12-18 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Design of a Small Molecule CD4-Antagonist with Broad Spectrum Anti-HIV-1 Activity.
J.Med.Chem., 58, 2015
|
|
6FXC
 
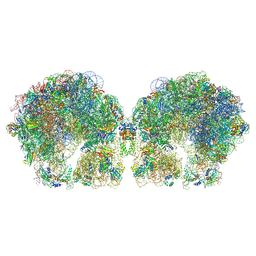 | | The cryo-EM structure of hibernating 100S ribosome dimer from pathogenic Staphylococcus aureus | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Matzov, D, Aibara, S, Zimmerman, E, Bashan, A, Kidmose, R, Amunts, A, Yonath, A. | | Deposit date: | 2018-03-08 | | Release date: | 2018-03-21 | | Last modified: | 2018-11-21 | | Method: | ELECTRON MICROSCOPY (6.76 Å) | | Cite: | The cryo-EM structure of hibernating 100S ribosome dimer from pathogenic Staphylococcus aureus.
Nat Commun, 8, 2017
|
|
5T7V
 
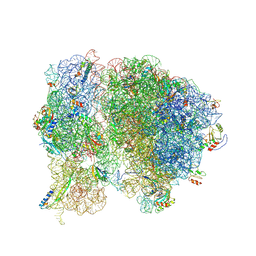 | | Methicillin Resistant, Linezolid resistant Staphylococcus aureus 70S ribosome (delta S145 uL3) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Belousoff, M.J, Lithgow, T, Eyal, Z, Yonath, A, Radjainia, M. | | Deposit date: | 2016-09-06 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis for Linezolid Binding Site Rearrangement in theStaphylococcus aureusRibosome.
MBio, 8, 2017
|
|
6GDS
 
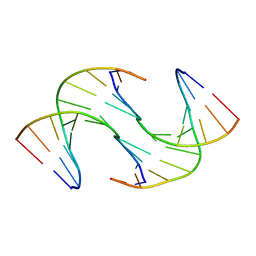 | | Holliday Junctions formed from Telomeric DNA | | Descriptor: | Telomeric DNA (5' CTAACCCTAA) 10mer, Telomeric DNA (5'-TTAGGGTTAG)-3') 10mer | | Authors: | Parkinson, G.N, Haider, S, Li, P, Khiali, S, Munnur, D, Ramanathan, A. | | Deposit date: | 2018-04-24 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Holliday Junctions Formed from Human Telomeric DNA.
J. Am. Chem. Soc., 140, 2018
|
|
6GDH
 
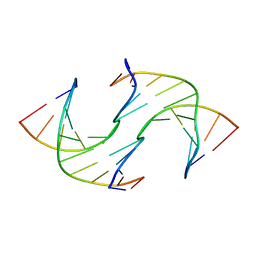 | | Holliday Junctions formed from Telomeric DNA | | Descriptor: | DNA (5'-D(*CP*TP*AP*AP*CP*CP*CP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*AP*GP*GP*GP*TP*TP*AP*G)-3') | | Authors: | Parkinson, G.N, Haider, S, Li, P, Khiali, S, Munnur, D, Ramanathan, A. | | Deposit date: | 2018-04-23 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Holliday Junctions Formed from Human Telomeric DNA.
J. Am. Chem. Soc., 140, 2018
|
|
4U67
 
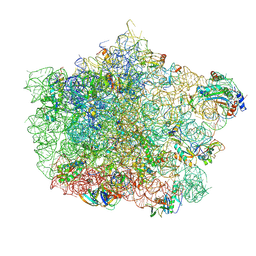 | | Crystal structure of the large ribosomal subunit (50S) of Deinococcus radiodurans containing a three residue insertion in L22 | | Descriptor: | 23s RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Wekselman, I, Zimmerman, E, Rozenberg, H, Bashan, A, Yonath, A. | | Deposit date: | 2014-07-28 | | Release date: | 2015-08-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | The Ribosomal Protein uL22 Modulates the Shape of the Protein Exit Tunnel.
Structure, 25, 2017
|
|
6HMA
 
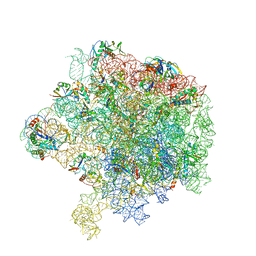 | | Improved model derived from cryo-EM map of Staphylococcus aureus large ribosomal subunit | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Eyal, Z, Cimicata, G, Matzov, D, Fox, T, de Val, N, Zimmerman, E, Bashan, A, Yonath, A. | | Deposit date: | 2018-09-12 | | Release date: | 2018-11-14 | | Last modified: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Improved model derived from cryo-EM map of Staphylococcus aureus large ribosomal subunit
To Be Published
|
|
5TCU
 
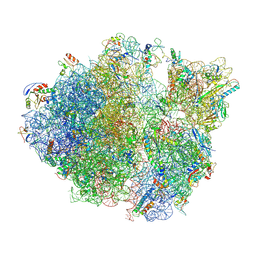 | | Methicillin sensitive Staphylococcus aureus 70S ribosome | | Descriptor: | 16S RRNA, 23S RRNA, 30S ribosomal protein S10, ... | | Authors: | Eyal, Z, Ahmed, T, Belousoff, N, Mishra, S, Matzov, D, Bashan, A, Zimmerman, E, Lithgow, T, Bhushan, S, Yonath, A. | | Deposit date: | 2016-09-15 | | Release date: | 2017-05-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis for Linezolid Binding Site Rearrangement in the Staphylococcus aureus Ribosome.
MBio, 8, 2017
|
|
6AZ1
 
 | | Cryo-EM structure of the small subunit of Leishmania ribosome bound to paromomycin | | Descriptor: | E-site tRNA, LACK1, MAGNESIUM ION, ... | | Authors: | Shalev-Benami, M, Zhang, Y, Rozenberg, H, Matzov, D, Zimmerman, E, Bashan, A, Jaffe, C.L, Yonath, A, Skiniotis, G. | | Deposit date: | 2017-09-09 | | Release date: | 2017-12-06 | | Last modified: | 2019-07-03 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Atomic resolution snapshot of Leishmania ribosome inhibition by the aminoglycoside paromomycin.
Nat Commun, 8, 2017
|
|
6AZ3
 
 | | Cryo-EM structure of of the large subunit of Leishmania ribosome bound to paromomycin | | Descriptor: | 60S ribosomal protein L10, putative, 60S ribosomal protein L11 (L5, ... | | Authors: | Shalev-Benami, M, Zhang, Y, Rozenberg, H, Nobe, Y, Taoka, M, Matzov, D, Zimmerman, E, Bashan, A, Isobe, T, Jaffe, C.L, Yonath, A, Skiniotis, G. | | Deposit date: | 2017-09-09 | | Release date: | 2017-12-06 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Atomic resolution snapshot of Leishmania ribosome inhibition by the aminoglycoside paromomycin.
Nat Commun, 8, 2017
|
|
5U6E
 
 | | Crystal structure of clade A/E HIV-1 gp120 core in complex with NBD-14010 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, N-{(1S)-2-amino-1-[5-(hydroxymethyl)-4-methyl-1,3-thiazol-2-yl]ethyl}-5-(4-chloro-3-fluorophenyl)-1H-pyrrole-2-carboxamide, ... | | Authors: | Kwon, Y.D, Debnath, A.K, Kwong, P.D. | | Deposit date: | 2016-12-07 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Synthesis, Antiviral Potency, in Vitro ADMET, and X-ray Structure of Potent CD4 Mimics as Entry Inhibitors That Target the Phe43 Cavity of HIV-1 gp120.
J. Med. Chem., 60, 2017
|
|
2C45
 
 | |
5UJC
 
 | | Crystal structure of a C.elegans B12-trafficking protein CblC, a human MMACHC homologue | | Descriptor: | CO-METHYLCOBALAMIN, D(-)-TARTARIC ACID, GLYCEROL, ... | | Authors: | Li, Z, Shanmuganathan, A, Ruetz, M, Yamada, K, Lesniak, N.A, Krautler, B, Brunold, T.C, Banerjee, R, Koutmos, M. | | Deposit date: | 2017-01-17 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Coordination chemistry controls the thiol oxidase activity of the B12-trafficking protein CblC.
J. Biol. Chem., 292, 2017
|
|
5HL7
 
 | | The crystal structure of the large ribosomal subunit of Staphylococcus aureus in complex with lefamulin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 23S ribosomal RNA, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eyal, Z, Matzov, D, Krupkin, M, Rozenberg, H, Zimmerman, E, Bashan, A, Yonath, A. | | Deposit date: | 2016-01-14 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | A novel pleuromutilin antibacterial compound, its binding mode and selectivity mechanism.
Sci Rep, 6, 2016
|
|
5TL8
 
 | | Naegleria fowleri CYP51-posaconazole complex | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, POSACONAZOLE, ... | | Authors: | Podust, L.M, Jennings, G, Calvet-Alvarez, C, Debnath, A. | | Deposit date: | 2016-10-10 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure of the Naegleria fowleri CYP51 at 1.7 Angstroms resolution
To be published
|
|
7DSU
 
 | | Structure of Mod subunit of the Type III restriction-modification enzyme Mbo45V | | Descriptor: | 1,2-ETHANEDIOL, Mbo45V, SINEFUNGIN | | Authors: | Ahmed, I, Chouhan, O.P, Gopinath, A, Morgan, R.D, Bhagat, K, Singh, A, Saikrishnan, K. | | Deposit date: | 2021-01-02 | | Release date: | 2022-01-05 | | Last modified: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of Mod subunit of the Type III restriction-modification enzyme Mbo45V
To Be Published
|
|
1FKA
 
 | | STRUCTURE OF FUNCTIONALLY ACTIVATED SMALL RIBOSOMAL SUBUNIT AT 3.3 A RESOLUTION | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Schluenzen, F, Tocilj, A, Zarivach, R, Harms, J, Gluehmann, M, Janell, D, Bashan, A, Bartels, H, Agmon, I, Franceschi, F, Yonath, A. | | Deposit date: | 2000-08-09 | | Release date: | 2000-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution.
Cell(Cambridge,Mass.), 102, 2000
|
|
1NJP
 
 | | The crystal structure of the 50S Large ribosomal subunit from Deinococcus radiodurans complexed with a tRNA acceptor stem mimic (ASM) | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L16, GENERAL STRESS PROTEIN CTC, ... | | Authors: | Bashan, A, Agmon, I, Zarivatch, R, Schluenzen, F, Harms, J.M, Berisio, R, Bartels, H, Hansen, H.A, Yonath, A. | | Deposit date: | 2003-01-02 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of the ribosomal machinery for Peptide bond formation,
translocation, and nascent chain progression
Mol.Cell, 11, 2003
|
|
1NKW
 
 | | Crystal Structure Of The Large Ribosomal Subunit From Deinococcus Radiodurans | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | Authors: | Harms, J.M, Schluenzen, F, Zarivach, R, Bashan, A, Gat, S, Agmon, I, Bartels, H, Franceschi, F, Yonath, A. | | Deposit date: | 2003-01-05 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | High resolution structure of the large ribosomal subunit from a mesophilic eubacterium
Cell(Cambridge,Mass.), 107, 2001
|
|
1NJM
 
 | | The crystal structure of the 50S Large ribosomal subunit from Deinococcus radiodurans complexed with a tRNA acceptor stem mimic (ASM) and the antibiotic sparsomycin | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L16, GENERAL STRESS PROTEIN CTC, ... | | Authors: | Bashan, A, Agmon, I, Zarivatch, R, Schluenzen, F, Harms, J.M, Berisio, R, Bartels, H, Hansen, H.A, Yonath, A. | | Deposit date: | 2003-01-02 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis of the ribosomal machinery for Peptide bond formation,
translocation, and nascent chain progression
Mol.Cell, 11, 2003
|
|
1NJO
 
 | | The crystal structure of the 50S Large ribosomal subunit from Deinococcus radiodurans complexed with a short substrate analog ACCPuromycin (ACCP) | | Descriptor: | 23S ribosomal RNA, RNA ACC(Puromycin) | | Authors: | Bashan, A, Agmon, I, Zarivatch, R, Schluenzen, F, Harms, J.M, Berisio, R, Bartels, H, Hansen, H.A, Yonath, A. | | Deposit date: | 2003-01-02 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis of the ribosomal machinery for Peptide bond formation,
translocation, and nascent chain progression
Mol.Cell, 11, 2003
|
|
1I94
 
 | | CRYSTAL STRUCTURES OF THE SMALL RIBOSOMAL SUBUNIT WITH TETRACYCLINE, EDEINE AND IF3 | | Descriptor: | 16S RRNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Pioletti, M, Schluenzen, F, Harms, J, Zarivach, R, Gluehmann, M, Avila, H, Bartels, H, Jacobi, C, Hartsch, T, Yonath, A, Franceschi, F. | | Deposit date: | 2001-03-18 | | Release date: | 2001-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3.
EMBO J., 20, 2001
|
|
1I97
 
 | | CRYSTAL STRUCTURE OF THE 30S RIBOSOMAL SUBUNIT FROM THERMUS THERMOPHILUS IN COMPLEX WITH TETRACYCLINE | | Descriptor: | 16S RRNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Pioletti, M, Schluenzen, F, Harms, J, Zarivach, R, Gluehmann, M, Avila, H, Bartels, H, Jacobi, C, Hartsch, T, Yonath, A, Franceschi, F. | | Deposit date: | 2001-03-18 | | Release date: | 2001-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3.
EMBO J., 20, 2001
|
|
1I96
 
 | | CRYSTAL STRUCTURE OF THE 30S RIBOSOMAL SUBUNIT FROM THERMUS THERMOPHILUS IN COMPLEX WITH THE TRANSLATION INITIATION FACTOR IF3 (C-TERMINAL DOMAIN) | | Descriptor: | 16S RRNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Pioletti, M, Schluenzen, F, Harms, J, Zarivach, R, Gluehmann, M, Avila, H, Bartels, H, Jacobi, C, Hartsch, T, Yonath, A, Franceschi, F. | | Deposit date: | 2001-03-18 | | Release date: | 2001-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3.
EMBO J., 20, 2001
|
|
1I95
 
 | | CRYSTAL STRUCTURE OF THE 30S RIBOSOMAL SUBUNIT FROM THERMUS THERMOPHILUS IN COMPLEX WITH EDEINE | | Descriptor: | 16S RRNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Pioletti, M, Schluenzen, F, Harms, J, Zarivach, R, Gluehmann, M, Avila, H, Bartels, H, Jacobi, C, Hartsch, T, Yonath, A, Franceschi, F. | | Deposit date: | 2001-03-18 | | Release date: | 2001-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3.
EMBO J., 20, 2001
|
|
