8THO
 
 | |
8U7J
 
 | | Crystal Structure of Staphylococcus aureus PLP synthase complex | | Descriptor: | GLUTAMINE, PHOSPHATE ION, Pyridoxal 5'-phosphate synthase subunit PdxS, ... | | Authors: | Barra, A.L.C, Brognaro, H, Betzel, C, Nascimento, A.S. | | Deposit date: | 2023-09-15 | | Release date: | 2024-07-24 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structure and dynamics of the staphylococcal pyridoxal 5-phosphate synthase complex reveal transient interactions at the enzyme interface.
J.Biol.Chem., 300, 2024
|
|
7DAA
 
 | | Crystal structure of basigin complexed with anti-basigin Fab fragment | | Descriptor: | CADMIUM ION, Heavy chain of antibody Fab fragment, Isoform 2 of Basigin, ... | | Authors: | Sakuragi, T, Kanai, R, Narita, H, Onishi, E, Miyazaki, T, Baba, T, Nakagawa, A, Toyoshima, C, Nagata, S. | | Deposit date: | 2020-10-16 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | The tertiary structure of the human Xkr8-Basigin complex that scrambles phospholipids at plasma membranes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7DCE
 
 | | Cryo-EM structure of human XKR8-basigin complex bound to Fab fragment | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Heavy chain of Fab fragment, Isoform 2 of Basigin, ... | | Authors: | Sakuragi, T, Kanai, R, Tsutsumi, A, Narita, H, Onishi, E, Miyazaki, T, Baba, T, Nakagawa, A, Kikkawa, M, Toyoshima, C, Nagata, S. | | Deposit date: | 2020-10-26 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The tertiary structure of the human Xkr8-Basigin complex that scrambles phospholipids at plasma membranes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7D9Z
 
 | | Crystal structure of anti-basigin Fab fragment | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Heavy chain of antibody Fab fragment, ... | | Authors: | Sakuragi, T, Kanai, R, Narita, H, Onishi, E, Miyazaki, T, Baba, T, Nakagawa, A, Toyoshima, C, Nagata, S. | | Deposit date: | 2020-10-14 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.123 Å) | | Cite: | The tertiary structure of the human Xkr8-Basigin complex that scrambles phospholipids at plasma membranes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
4ZXK
 
 | |
5A3G
 
 | | Structure of herpesvirus nuclear egress complex subunit M50 | | Descriptor: | M50 | | Authors: | Leigh, K.E, Boeszoermenyi, A, Mansueto, M.S, Sharma, M, Filman, D.J, Coen, D.M, Wagner, G, Hogle, J.M, Arthanari, H. | | Deposit date: | 2015-06-01 | | Release date: | 2015-07-15 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure of a Herpesvirus Nuclear Egress Complex Subunit Reveals an Interaction Groove that is Essential for Viral Replication
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5BXV
 
 | | eIF4E complex | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4E, Eukaryotic translation initiation factor 4E-binding protein 1 | | Authors: | Sekiyama, N, Arthanari, H, Papdopoulos, E, Rodriguez-Mias, R.A, Wagner, G, Leger-Abraham, M. | | Deposit date: | 2015-06-09 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular mechanism of the dual activity of 4EGI-1: Dissociating eIF4G from eIF4E but stabilizing the binding of unphosphorylated 4E-BP1.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5B22
 
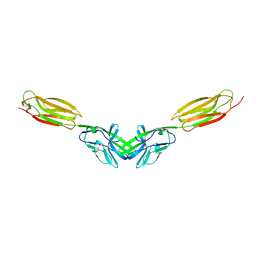 | | Dimer structure of murine Nectin-3 D1D2 | | Descriptor: | Nectin-3, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Takebe, K, Sangawa, T, Katsutani, T, Narita, H, Suzuki, M. | | Deposit date: | 2015-12-28 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Dimer structure of murine Nectin-3 D1D2
To Be Published
|
|
5IJ4
 
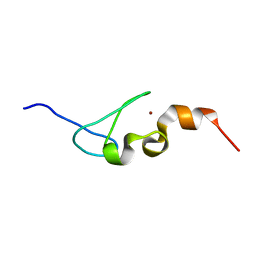 | | Solution structure of AN1-type zinc finger domain from Cuz1 (Cdc48 associated ubiquitin-like/zinc-finger protein-1) | | Descriptor: | CDC48-associated ubiquitin-like/zinc finger protein 1, ZINC ION | | Authors: | Sun, Z.-Y.J, Hanna, J, Wagner, G, Bhanu, M.K, Allan, M, Arthanari, H. | | Deposit date: | 2016-03-01 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Cuz1 AN1 Zinc Finger Domain: An Exposed LDFLP Motif Defines a Subfamily of AN1 Proteins.
Plos One, 11, 2016
|
|
3LKF
 
 | | LEUKOCIDIN F (HLGB) FROM STAPHYLOCOCCUS AUREUS WITH PHOSPHOCHOLINE BOUND | | Descriptor: | LEUKOCIDIN F SUBUNIT, PHOSPHOCHOLINE | | Authors: | Olson, R, Nariya, H, Yokota, K, Kamio, Y, Gouaux, J.E. | | Deposit date: | 1998-07-28 | | Release date: | 1999-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of staphylococcal LukF delineates conformational changes accompanying formation of a transmembrane channel.
Nat.Struct.Biol., 6, 1999
|
|
3OYP
 
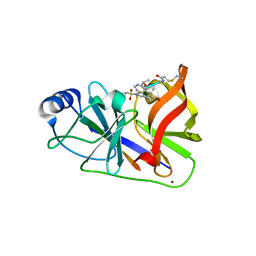 | | HCV NS3/4A in complex with ligand 3 | | Descriptor: | Non-structural protein 4A, Peptidomimetic inhibitor, Serine protease NS3, ... | | Authors: | Hagel, M, Niu, D, St.Martin, T, Sheets, M.P, Qiao, L, Bernard, H, Karp, R.M, Zhu, Z, Labenski, M.T, Chaturvedi, P.C, Nacht, M, Westlin, W.F, Petter, R.C, Singh, J. | | Deposit date: | 2010-09-23 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Selective irreversible inhibition of a protease by targeting a noncatalytic cysteine.
Nat.Chem.Biol., 7, 2011
|
|
3VQF
 
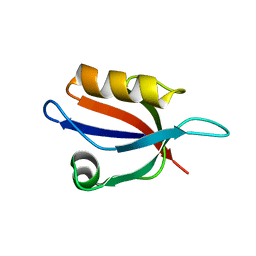 | | Crystal Structure Analysis of the PDZ Domain Derived from the Tight Junction Regulating Protein | | Descriptor: | E3 ubiquitin-protein ligase LNX | | Authors: | Akiyoshi, Y, Hamada, D, Goda, N, Tenno, T, Narita, H, Nakagawa, A, Furuse, M, Suzuki, M, Hiroaki, H. | | Deposit date: | 2012-03-22 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Structural basis for down regulation of tight junction by PDZ-domain containing E3-Ubiquitin ligase
To be Published
|
|
3VQG
 
 | | Crystal Structure Analysis of the PDZ Domain Derived from the Tight Junction Regulating Protein | | Descriptor: | C-terminal peptide from Immunoglobulin superfamily member 5, E3 ubiquitin-protein ligase LNX, SULFATE ION | | Authors: | Akiyoshi, Y, Hamada, D, Goda, N, Tenno, T, Narita, H, Nakagawa, A, Furuse, M, Suzuki, M, Hiroaki, H. | | Deposit date: | 2012-03-23 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis for down regulation of tight junction by PDZ-domain containing E3-Ubiquitin ligase
To be Published
|
|
3WIH
 
 | | Crystal structure of the third fibronectin domain (Fn3) of human ROBO1 in complex with the Fab fragment of murine monoclonal antibody B2212A. | | Descriptor: | GLYCEROL, Roundabout homolog 1, anti-human ROBO1 antibody B2212A Fab heavy chain, ... | | Authors: | Nakayama, T, Mizohata, E, Yamashita, T, Nagatoishi, M, Iwanari, H, Mochizuki, Y, Kado, Y, Yokota, Y, Sato, R, Tsumoto, K, Fujitani, H, Kodama, T, Hamakubo, T, Inoue, T. | | Deposit date: | 2013-09-12 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structural features of interfacial tyrosine residue in ROBO1 fibronectin domain-antibody complex: Crystallographic, thermodynamic, and molecular dynamic analyses
Protein Sci., 24, 2015
|
|
5WB5
 
 | | Leishmania IF4E-1 bound to Leishmania 4E-IP1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Putative eukaryotic translation initiation factor eIF-4E, Uncharacterized protein | | Authors: | Leger-Abraham, M, Meleppattu, S, Arthanari, H, Zinoviev, A, Boeszoermenyi, A, Wagner, G, Shapira, M. | | Deposit date: | 2017-06-27 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for LeishIF4E-1 modulation by an interacting protein in the human parasite Leishmania major.
Nucleic Acids Res., 46, 2018
|
|
4D7X
 
 | |
4C1W
 
 | | Carbohydrate binding domain from Streptococcus pneumoniae NanA sialidase complexed with 3'-sialyllactose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, NEURAMINIDASE | | Authors: | Yang, L, Connaris, H, Potter, J.A, Taylor, G.L. | | Deposit date: | 2013-08-14 | | Release date: | 2014-08-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Prevention of Influenza by Targeting Host Receptors Using Engineered Proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WII
 
 | | Crystal structure of the Fab fragment of B2212A, a murine monoclonal antibody specific for the third fibronectin domain (Fn3) of human ROBO1. | | Descriptor: | anti-human ROBO1 antibody B2212A Fab heavy chain, anti-human ROBO1 antibody B2212A Fab light chain | | Authors: | Nakayama, T, Mizohata, E, Yamashita, T, Nagatoishi, S, Nakakido, M, Iwanari, H, Mochizuki, Y, Kado, Y, Yokota, Y, Sato, R, Tsumoto, K, Fujitani, H, Kodama, T, Hamakubo, T, Inoue, T. | | Deposit date: | 2013-09-12 | | Release date: | 2015-01-21 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural features of interfacial tyrosine residue in ROBO1 fibronectin domain-antibody complex: Crystallographic, thermodynamic, and molecular dynamic analyses
Protein Sci., 24, 2015
|
|
3VG9
 
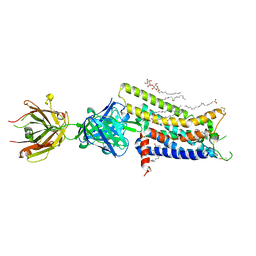 | | Crystal structure of human adenosine A2A receptor with an allosteric inverse-agonist antibody at 2.7 A resolution | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Hino, T, Arakawa, T, Iwanari, H, Yurugi-Kobayashi, T, Ikeda-Suno, C, Nakada-Nakura, Y, Kusano-Arai, O, Weyand, S, Shimamura, T, Nomura, N, Cameron, A.D, Kobayashi, T, Hamakubo, T, Iwata, S, Murata, T. | | Deposit date: | 2011-08-04 | | Release date: | 2012-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | G-protein-coupled receptor inactivation by an allosteric inverse-agonist antibody
Nature, 482, 2012
|
|
3VGA
 
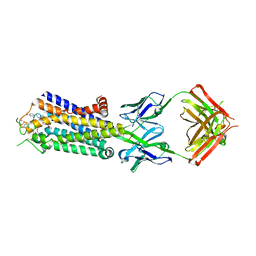 | | Crystal structure of human adenosine A2A receptor with an allosteric inverse-agonist antibody at 3.1 A resolution | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a, antibody fab fragment heavy chain, ... | | Authors: | Hino, T, Arakawa, T, Iwanari, H, Yurugi-Kobayashi, T, Ikeda-Suno, C, Nakada-Nakura, Y, Kusano-Arai, O, Weyand, S, Shimamura, T, Nomura, N, Cameron, A.D, Kobayashi, T, Hamakubo, T, Iwata, S, Murata, T. | | Deposit date: | 2011-08-04 | | Release date: | 2012-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | G-protein-coupled receptor inactivation by an allosteric inverse-agonist antibody
Nature, 482, 2012
|
|
3ASQ
 
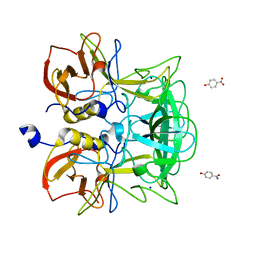 | | Crystal structure of P domain from Norovirus Funabashi258 stain in the complex with H-antigen | | Descriptor: | Capsid protein, P-NITROPHENOL, SODIUM ION, ... | | Authors: | Kubota, T, Kumagai, A, Itoh, H, Furukawa, S, Narimatsu, H, Wakita, T, Ishii, K, Takeda, N, Someya, Y, Shirato, H. | | Deposit date: | 2010-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of Lewis antigens by genogroup I norovirus
J.Virol., 86, 2012
|
|
3ASR
 
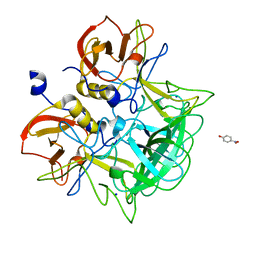 | | Crystal structure of P domain from Norovirus Funabashi258 stain in the complex with Lewis-a | | Descriptor: | Capsid protein, P-NITROPHENOL, SODIUM ION, ... | | Authors: | Kubota, T, Kumagai, A, Itoh, H, Furukawa, S, Narimatsu, H, Wakita, T, Ishii, K, Takeda, N, Someya, Y, Shirato, H. | | Deposit date: | 2010-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of Lewis antigens by genogroup I norovirus
J.Virol., 86, 2012
|
|
3ASS
 
 | | Crystal structure of P domain from Norovirus Funabashi258 stain in the complex with Lewis-b | | Descriptor: | Capsid protein, P-NITROPHENOL, SODIUM ION, ... | | Authors: | Kubota, T, Kumagai, A, Itoh, H, Furukawa, S, Narimatsu, H, Wakita, T, Ishii, K, Takeda, N, Someya, Y, Shirato, H. | | Deposit date: | 2010-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of Lewis antigens by genogroup I norovirus
J.Virol., 86, 2012
|
|
3ASP
 
 | | Crystal structure of P domain from Norovirus Funabashi258 stain in the complex with A-antigen | | Descriptor: | Capsid protein, SODIUM ION, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kubota, T, Kumagai, A, Itoh, H, Furukawa, S, Narimatsu, H, Wakita, T, Ishii, K, Takeda, N, Someya, Y, Shirato, H. | | Deposit date: | 2010-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of Lewis antigens by genogroup I norovirus
J.Virol., 86, 2012
|
|
