5CIZ
 
 | |
1ZYR
 
 | | Structure of Thermus thermophilus RNA polymerase holoenzyme in complex with the antibiotic streptolydigin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase omega chain, ... | | Authors: | Tuske, S, Sarafianos, S.G, Wang, X, Hudson, B, Sineva, E, Mukhopadhyay, J, Birktoft, J.J, Leroy, O, Ismail, S, Clark, A.D, Dharia, C, Napoli, A, Laptenko, O, Lee, J, Borukhov, S, Ebright, R.H, Arnold, E. | | Deposit date: | 2005-06-10 | | Release date: | 2005-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Inhibition of bacterial RNA polymerase by streptolydigin: stabilization of a straight-bridge-helix active-center conformation.
Cell(Cambridge,Mass.), 122, 2005
|
|
2CW0
 
 | | Crystal structure of Thermus thermophilus RNA polymerase holoenzyme at 3.3 angstroms resolution | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Tuske, S, Sarafianos, S.G, Wang, X, Hudson, B, Sineva, E, Mukhopadhyay, J, Birktoft, J.J, Leroy, O, Ismail, S, Clark Jr, A.D, Dharia, C, Napoli, A, Laptenko, O, Lee, J, Borukhov, S, Ebright, R.H, Arnold, E. | | Deposit date: | 2005-06-15 | | Release date: | 2005-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Inhibition of bacterial RNA polymerase by streptolydigin: stabilization of a straight-bridge-helix active-center conformation
Cell(Cambridge,Mass.), 122, 2005
|
|
1ZRD
 
 | | 4 crystal structures of CAP-DNA with all base-pair substitutions at position 6, CAP-[6A;17T]ICAP38 DNA | | Descriptor: | 5'-D(*AP*TP*TP*TP*CP*GP*AP*AP*AP*AP*AP*TP*GP*AP*GP*AP*T)-3', 5'-D(*CP*TP*AP*GP*AP*TP*CP*TP*CP*AP*TP*TP*TP*TP*TP*CP*GP*AP*AP*AP*T)-3', ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ... | | Authors: | Berman, H.M, Napoli, A.A. | | Deposit date: | 2005-05-19 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Indirect readout of DNA sequence at the primary-kink site in the CAP-DNA complex: recognition of pyrimidine-purine and purine-purine steps.
J.Mol.Biol., 357, 2006
|
|
1ZRF
 
 | | 4 crystal structures of CAP-DNA with all base-pair substitutions at position 6, CAP-[6C;17G]ICAP38 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 5'-D(*AP*TP*TP*TP*CP*GP*AP*AP*AP*AP*AP*TP*GP*CP*GP*AP*T)-3', 5'-D(*CP*TP*AP*GP*AP*TP*CP*GP*CP*AP*TP*TP*TP*TP*TP*CP*GP*AP*AP*AP*T)-3', ... | | Authors: | Berman, H.M, Napoli, A.A. | | Deposit date: | 2005-05-19 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Indirect readout of DNA sequence at the primary-kink site in the CAP-DNA complex: recognition of pyrimidine-purine and purine-purine steps.
J.Mol.Biol., 357, 2006
|
|
1ZRC
 
 | | 4 Crystal structures of CAP-DNA with all base-pair substitutions at position 6, CAP-ICAP38 DNA | | Descriptor: | 5'-D(*AP*TP*TP*TP*CP*GP*AP*AP*AP*AP*AP*TP*GP*TP*GP*AP*T)-3', 5'-D(*CP*TP*AP*GP*AP*TP*CP*AP*CP*AP*TP*TP*TP*TP*TP*CP*GP*AP*AP*AP*T)-3', ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ... | | Authors: | Berman, H.M, Napoli, A.A. | | Deposit date: | 2005-05-19 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Indirect readout of DNA sequence at the primary-kink site in the CAP-DNA complex: recognition of pyrimidine-purine and purine-purine steps.
J.Mol.Biol., 357, 2006
|
|
1ZRE
 
 | | 4 crystal structures of CAP-DNA with all base-pair substitutions at position 6, CAP-[6G;17C]ICAP38 DNA | | Descriptor: | 5'-D(*AP*TP*TP*TP*CP*GP*AP*AP*AP*AP*AP*TP*GP*GP*GP*AP*T)-3', 5'-D(*CP*TP*AP*GP*AP*TP*CP*CP*CP*AP*TP*TP*TP*TP*TP*CP*GP*AP*AP*AP*T)-3', ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ... | | Authors: | Berman, H.M, Napoli, A.A. | | Deposit date: | 2005-05-19 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Indirect readout of DNA sequence at the primary-kink site in the CAP-DNA complex: recognition of pyrimidine-purine and purine-purine steps.
J.Mol.Biol., 357, 2006
|
|
2B1C
 
 | |
2B1B
 
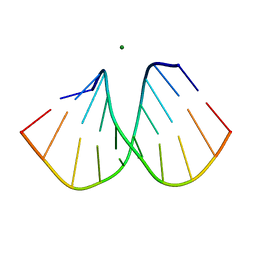 | |
2B1D
 
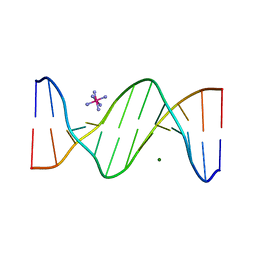 | |
