6GV1
 
 | | Crystal structure of E.coli Multidrug/H+ antiporter MdfA in outward open conformation with bound Fab fragment | | 分子名称: | Fab fragment YN1074 heavy chain, Fab fragment YN1074 light chain, Major Facilitator Superfamily multidrug/H+ antiporter MdfA from E.coli, ... | | 著者 | Nagarathinam, K, Parthier, C, Stubbs, M.T, Tanabe, M. | | 登録日 | 2018-06-20 | | 公開日 | 2018-10-03 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Outward open conformation of a Major Facilitator Superfamily multidrug/H+antiporter provides insights into switching mechanism.
Nat Commun, 9, 2018
|
|
6IAK
 
 | | The crystal structure of the chicken CREB3 bZIP | | 分子名称: | Uncharacterized protein | | 著者 | Sabaratnam, K, Renner, M. | | 登録日 | 2018-11-26 | | 公開日 | 2019-12-11 | | 最終更新日 | 2020-06-24 | | 実験手法 | X-RAY DIFFRACTION (3.95 Å) | | 主引用文献 | Insights from the crystal structure of the chicken CREB3 bZIP suggest that members of the CREB3 subfamily transcription factors may be activated in response to oxidative stress.
Protein Sci., 28, 2019
|
|
5L6Q
 
 | | Refolded AL protein from cardiac amyloidosis | | 分子名称: | CARBONATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Annamalai, K, Liberta, F, Vielberg, M.-T, Lilie, H, Guehrs, K.-H, Schierhorn, A, Koehler, R, Schmidt, A, Haupt, C, Hegenbart, O, Schoenland, S, Groll, M, Faendrich, M. | | 登録日 | 2016-05-31 | | 公開日 | 2017-05-31 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Common Fibril Structures Imply Systemically Conserved Protein Misfolding Pathways In Vivo.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6LNG
 
 | | Rapid crystallization of streptavidin using charged peptides | | 分子名称: | GLYCEROL, Streptavidin | | 著者 | Minamihata, K, Tsukamoto, K, Adachi, M, Shimizu, R, Mishina, M, Kuroki, R, Nagamune, T. | | 登録日 | 2019-12-30 | | 公開日 | 2020-03-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8000015 Å) | | 主引用文献 | Genetically fused charged peptides induce rapid crystallization of proteins.
Chem.Commun.(Camb.), 56, 2020
|
|
7VEW
 
 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in complex with unsaturated trigalacturonic acid | | 分子名称: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | 著者 | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | 登録日 | 2021-09-10 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VEQ
 
 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in an open conformation | | 分子名称: | GLYCEROL, SPH1118 | | 著者 | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | 登録日 | 2021-09-10 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.696 Å) | | 主引用文献 | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VET
 
 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in a closed conformation | | 分子名称: | SPH1118 | | 著者 | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | 登録日 | 2021-09-10 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VEV
 
 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in complex with MES | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, SPH1118 | | 著者 | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | 登録日 | 2021-09-10 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.498 Å) | | 主引用文献 | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VER
 
 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in a full open conformation | | 分子名称: | GLYCEROL, SPH1118 | | 著者 | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | 登録日 | 2021-09-10 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.699 Å) | | 主引用文献 | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VEU
 
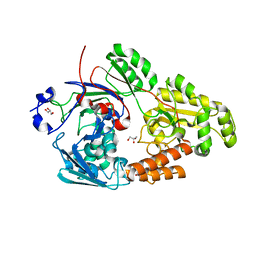 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in complex with galacturonic acid | | 分子名称: | GLYCEROL, SPH1118, alpha-D-galactopyranuronic acid | | 著者 | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | 登録日 | 2021-09-10 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.736 Å) | | 主引用文献 | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
1IKM
 
 | |
1IKL
 
 | |
1G91
 
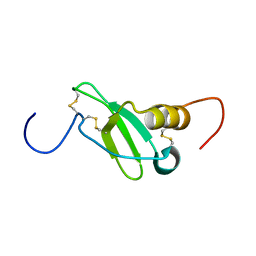 | | SOLUTION STRUCTURE OF MYELOID PROGENITOR INHIBITORY FACTOR-1 (MPIF-1) | | 分子名称: | MYELOID PROGENITOR INHIBITORY FACTOR-1 | | 著者 | Rajarathnam, K, Li, Y, Rohrer, T, Gentz, R. | | 登録日 | 2000-11-21 | | 公開日 | 2001-03-07 | | 最終更新日 | 2022-12-21 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and dynamics of myeloid progenitor inhibitory factor-1 (MPIF-1), a novel monomeric CC chemokine.
J.Biol.Chem., 276, 2001
|
|
7APU
 
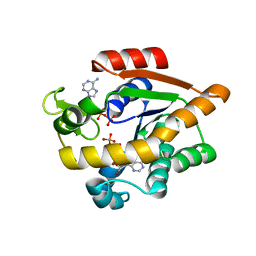 | | Structure of Adenylate kinase from Escherichia coli in complex with two ADP molecules refined at 1.36 A resolution. | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Adenylate kinase, SODIUM ION | | 著者 | Grundstom, C, Wolf-Watz, M, Nam, K, Sauer, U.H. | | 登録日 | 2020-10-19 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Dynamic Connection between Enzymatic Catalysis and Collective Protein Motions.
Biochemistry, 60, 2021
|
|
8Q2B
 
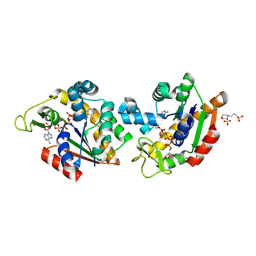 | | E. coli Adenylate Kinase variant D158A (AK D158A) showing significant changes to the stacking of catalytic arginine side chains | | 分子名称: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ... | | 著者 | Sauer, U.H, Wolf-Watz, M, Nam, K. | | 登録日 | 2023-08-01 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Elucidating Dynamics of Adenylate Kinase from Enzyme Opening to Ligand Release.
J.Chem.Inf.Model., 64, 2024
|
|
9IWQ
 
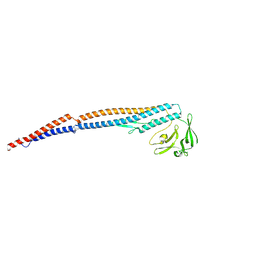 | | Salmonella enterica serovar Typhimurium FliC(G426A)delta(204-292) forming the L-type straight filament | | 分子名称: | Flagellin | | 著者 | Waraich, K, Makino, F, Miyata, T, Kinoshita, M, Minamino, T, Namba, K. | | 登録日 | 2024-07-25 | | 公開日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (2.08 Å) | | 主引用文献 | Salmonella enterica serovar Typhimurium FliC(G426A)delta(204-292) forming the L-type straight filament
To Be Published
|
|
8IBN
 
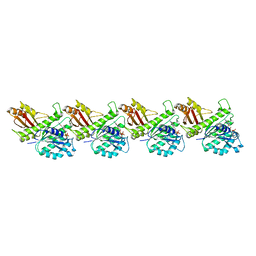 | | Cryo-EM structure of KpFtsZ single filament | | 分子名称: | Cell division protein FtsZ, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, POTASSIUM ION | | 著者 | Fujita, J, Amesaka, H, Yoshizawa, T, Kuroda, N, Kamimura, N, Hibino, K, Konishi, T, Kato, Y, Hara, M, Inoue, T, Namba, K, Tanaka, S, Matsumura, H. | | 登録日 | 2023-02-10 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.03 Å) | | 主引用文献 | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
2I0H
 
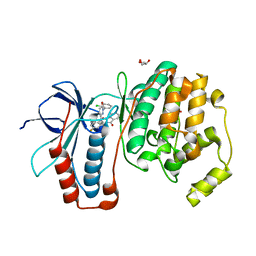 | | The structure of p38alpha in complex with an arylpyridazinone | | 分子名称: | 2-(3-{(2-CHLORO-4-FLUOROPHENYL)[1-(2-CHLOROPHENYL)-6-OXO-1,6-DIHYDROPYRIDAZIN-3-YL]AMINO}PROPYL)-1H-ISOINDOLE-1,3(2H)-DIONE, GLYCEROL, Mitogen-activated protein kinase 14 | | 著者 | Natarajan, S.R, Heller, S.T, Nam, K, Singh, S.B, Scapin, G, Patel, S, Thompson, J.E, Fitzgerald, C.E, O'Keefe, S.J. | | 登録日 | 2006-08-10 | | 公開日 | 2006-10-17 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | p38 MAP Kinase Inhibitors Part 6: 2-Arylpyridazin-3-ones as templates for inhibitor design.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
6JF2
 
 | |
1A05
 
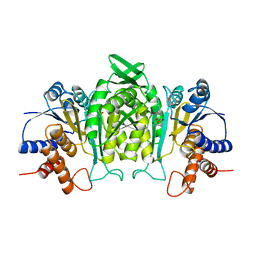 | | CRYSTAL STRUCTURE OF THE COMPLEX OF 3-ISOPROPYLMALATE DEHYDROGENASE FROM THIOBACILLUS FERROOXIDANS WITH 3-ISOPROPYLMALATE | | 分子名称: | 3-ISOPROPYLMALATE DEHYDROGENASE, 3-ISOPROPYLMALIC ACID, MAGNESIUM ION | | 著者 | Imada, K, Inagaki, K, Matsunami, H, Kawaguchi, H, Tanaka, H, Tanaka, N, Namba, K. | | 登録日 | 1997-12-09 | | 公開日 | 1998-06-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of 3-isopropylmalate dehydrogenase in complex with 3-isopropylmalate at 2.0 A resolution: the role of Glu88 in the unique substrate-recognition mechanism.
Structure, 6, 1998
|
|
3J6P
 
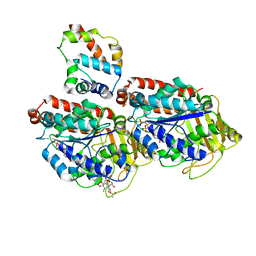 | | Pseudo-atomic model of dynein microtubule binding domain-tubulin complex based on a cryoEM map | | 分子名称: | Dynein heavy chain, cytoplasmic, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Uchimura, S, Fujii, T, Takazaki, H, Ayukawa, R, Nishikawa, Y, Minoura, I, Hachikubo, Y, Kurisu, G, Sutoh, K, Kon, T, Namba, K, Muto, E. | | 登録日 | 2014-03-20 | | 公開日 | 2014-12-31 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (8.2 Å) | | 主引用文献 | A flipped ion pair at the dynein-microtubule interface is critical for dynein motility and ATPase activation
J.Cell Biol., 208, 2015
|
|
7BWM
 
 | |
5H72
 
 | | Structure of the periplasmic domain of FliP | | 分子名称: | Flagellar biosynthetic protein FliP | | 著者 | Fukumura, T, Kawaguchi, T, Saijo-Hamano, Y, Namba, K, Minamino, T, Imada, K. | | 登録日 | 2016-11-16 | | 公開日 | 2017-08-02 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Assembly and stoichiometry of the core structure of the bacterial flagellar type III export gate complex
PLoS Biol., 15, 2017
|
|
5B0O
 
 | | Structure of the FliH-FliI complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Flagellar assembly protein FliH, Flagellum-specific ATP synthase | | 著者 | Imada, K, Uchida, Y, Kinoshita, M, Namba, K, Minamino, T. | | 登録日 | 2015-11-02 | | 公開日 | 2016-03-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Insight into the flagella type III export revealed by the complex structure of the type III ATPase and its regulator
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6XMN
 
 | |
