5GX2
 
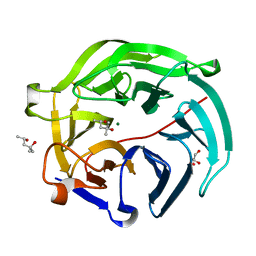 | | Luciferin-regenerating enzyme collected with serial synchrotron rotational crystallography with accumulated dose of 3.4 MGy (3rd measurement) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, Luciferin regenerating enzyme, ... | | Authors: | Hasegawa, K, Yamashita, K, Murai, T, Nuemket, N, Hirata, K, Ueno, G, Ago, H, Nakatsu, T, Kumasaka, T, Yamamoto, M. | | Deposit date: | 2016-09-15 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of a dose-limiting data collection strategy for serial synchrotron rotation crystallography
J Synchrotron Radiat, 24, 2017
|
|
5GX4
 
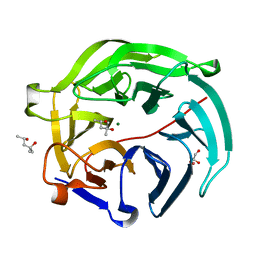 | | Luciferin-regenerating enzyme collected with serial synchrotron rotational crystallography with accumulated dose of 14 MGy (12th measurement) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, Luciferin regenerating enzyme, ... | | Authors: | Hasegawa, K, Yamashita, K, Murai, T, Nuemket, N, Hirata, K, Ueno, G, Ago, H, Nakatsu, T, Kumasaka, T, Yamamoto, M. | | Deposit date: | 2016-09-15 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of a dose-limiting data collection strategy for serial synchrotron rotation crystallography
J Synchrotron Radiat, 24, 2017
|
|
1K5C
 
 | | Endopolygalacturonase I from Stereum purpureum at 0.96 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ENDOPOLYGALACTURONASE, ... | | Authors: | Shimizu, T, Nakatsu, T, Miyairi, K, Okuno, T, Kato, H. | | Deposit date: | 2001-10-10 | | Release date: | 2002-06-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Active-site architecture of endopolygalacturonase I from Stereum purpureum revealed by crystal structures in native and ligand-bound forms at atomic resolution.
Biochemistry, 41, 2002
|
|
1AE1
 
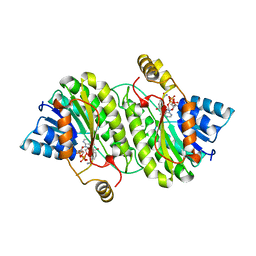 | | TROPINONE REDUCTASE-I COMPLEX WITH NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TROPINONE REDUCTASE-I | | Authors: | Nakajima, K, Yamashita, A, Akama, H, Nakatsu, T, Kato, H, Hashimoto, T, Oda, J, Yamada, Y. | | Deposit date: | 1997-10-23 | | Release date: | 1998-11-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of two tropinone reductases: different reaction stereospecificities in the same protein fold.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1AUN
 
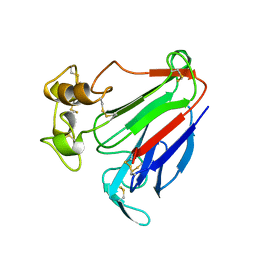 | | PATHOGENESIS-RELATED PROTEIN 5D FROM NICOTIANA TABACUM | | Descriptor: | PR-5D | | Authors: | Koiwa, H, Kato, H, Nakatsu, T, Oda, J, Yamada, Y, Sato, F. | | Deposit date: | 1997-08-29 | | Release date: | 1998-03-04 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of tobacco PR-5d protein at 1.8 A resolution reveals a conserved acidic cleft structure in antifungal thaumatin-like proteins.
J.Mol.Biol., 286, 1999
|
|
6A6M
 
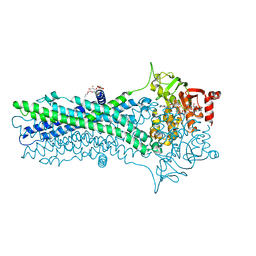 | | Crystal structure of an outward-open nucleotide-bound state of the eukaryotic ABC multidrug transporter CmABCB1 | | Descriptor: | ATP-binding cassette, sub-family B, member 1, ... | | Authors: | Kato, H, Nakatsu, T, Kodan, A. | | Deposit date: | 2018-06-28 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inward- and outward-facing X-ray crystal structures of homodimeric P-glycoprotein CmABCB1.
Nat Commun, 10, 2019
|
|
6A6N
 
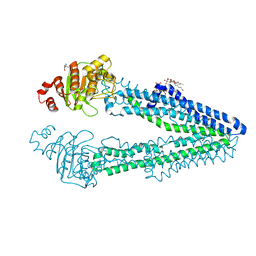 | | Crystal structure of an inward-open apo state of the eukaryotic ABC multidrug transporter CmABCB1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ATP-binding cassette, sub-family B, ... | | Authors: | Kato, H, Nakatsu, T, Kodan, A. | | Deposit date: | 2018-06-28 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Inward- and outward-facing X-ray crystal structures of homodimeric P-glycoprotein CmABCB1.
Nat Commun, 10, 2019
|
|
7DQV
 
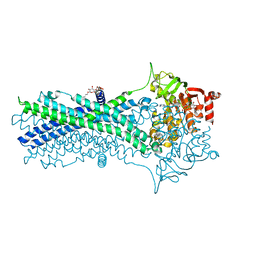 | | Crystal structure of a CmABCB1 mutant | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, MAGNESIUM ION, NITRATE ION, ... | | Authors: | Matsuoka, K, Nakatsu, T, Kato, H. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of the CmABCB1 G132V mutant, which favors the outward-facing state, reveals the mechanism of the pivotal joint between TM1 and TM3.
Protein Sci., 30, 2021
|
|
3ED1
 
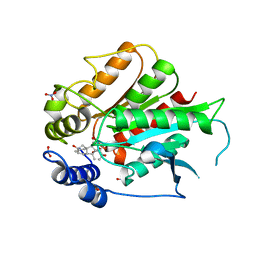 | | Crystal Structure of Rice GID1 complexed with GA3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GIBBERELLIN A3, Gibberellin receptor GID1, ... | | Authors: | Shimada, A, Nakatsu, T, Ueguchi-Tanaka, M, Kato, H, Matsuoka, M. | | Deposit date: | 2008-09-02 | | Release date: | 2008-11-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for gibberellin recognition by its receptor GID1.
Nature, 456, 2008
|
|
1V4G
 
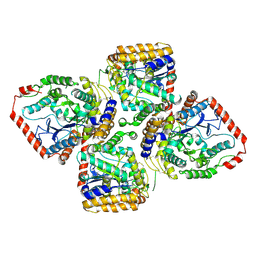 | | Crystal Structure of gamma-Glutamylcysteine Synthetase from Escherichia coli B | | Descriptor: | Glutamate--cysteine ligase | | Authors: | Hibi, T, Nii, H, Nakatsu, T, Kato, H, Hiratake, J, Oda, J. | | Deposit date: | 2003-11-13 | | Release date: | 2004-10-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of gamma-glutamylcysteine synthetase: insights into the mechanism of catalysis by a key enzyme for glutathione homeostasis
PROC.NATL.ACAD.SCI.USA, 101, 2004
|
|
1VA6
 
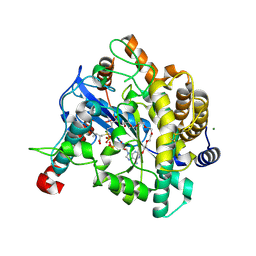 | | Crystal structure of Gamma-glutamylcysteine synthetase from Escherichia Coli B complexed with Transition-state analogue | | Descriptor: | (2S)-2-AMINO-4-[[(2R)-2-CARBOXYBUTYL](PHOSPHONO)SULFONIMIDOYL]BUTANOIC ACID, ADENOSINE-5'-DIPHOSPHATE, Glutamate--cysteine ligase, ... | | Authors: | Hibi, T, Nii, H, Nakatsu, T, Kato, H, Hiratake, J, Oda, J. | | Deposit date: | 2004-02-12 | | Release date: | 2004-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of gamma-glutamylcysteine synthetase: insights into the mechanism of catalysis by a key enzyme for glutathione homeostasis
PROC.NATL.ACAD.SCI.USA, 101, 2004
|
|
3EBL
 
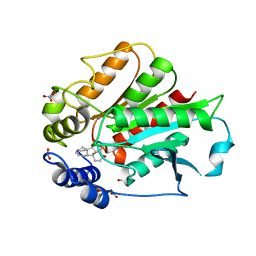 | | Crystal Structure of Rice GID1 complexed with GA4 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GIBBERELLIN A4, Gibberellin receptor GID1, ... | | Authors: | Shimada, A, Nakatsu, T, Ueguchi-Tanaka, M, Kato, H, Matsuoka, M. | | Deposit date: | 2008-08-28 | | Release date: | 2008-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for gibberellin recognition by its receptor GID1.
Nature, 456, 2008
|
|
1I2H
 
 | | CRYSTAL STRUCTURE ANALYSIS OF PSD-ZIP45(HOMER1C/VESL-1L)CONSERVED HOMER 1 DOMAIN | | Descriptor: | PSD-ZIP45(HOMER-1C/VESL-1L) | | Authors: | Irie, K, Nakatsu, T, Mitsuoka, K, Fujiyoshi, Y, Kato, H. | | Deposit date: | 2001-02-09 | | Release date: | 2002-05-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Homer 1 Family Conserved Region Reveals the Interaction Between the EVH1 Domain and
Own Proline-rich Motif
J.Mol.Biol., 318, 2002
|
|
1VBH
 
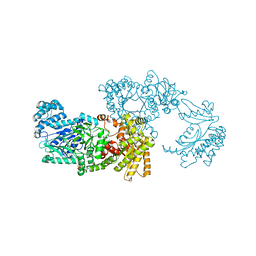 | | Pyruvate Phosphate Dikinase with bound Mg-PEP from Maize | | Descriptor: | MAGNESIUM ION, PHOSPHOENOLPYRUVATE, SULFATE ION, ... | | Authors: | Nakanishi, T, Nakatsu, T, Matsuoka, M, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-02-26 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of pyruvate phosphate dikinase from maize revealed an alternative conformation in the swiveling-domain motion
Biochemistry, 44, 2005
|
|
1VBG
 
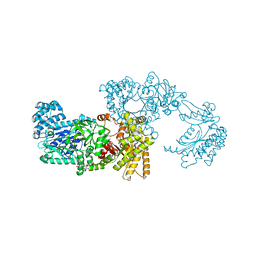 | | Pyruvate Phosphate Dikinase from Maize | | Descriptor: | MAGNESIUM ION, SULFATE ION, pyruvate,orthophosphate dikinase | | Authors: | Nakanishi, T, Nakatsu, T, Matsuoka, M, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-02-26 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of pyruvate phosphate dikinase from maize revealed an alternative conformation in the swiveling-domain motion
Biochemistry, 44, 2005
|
|
1V2B
 
 | | Crystal Structure of PsbP Protein in the Oxygen-Evolving Complex of Photosystem II from Higher Plants | | Descriptor: | 23-kDa polypeptide of photosystem II oxygen-evolving complex, SULFATE ION, alpha-D-glucopyranose | | Authors: | Ifuku, K, Nakatsu, T, Kato, H, Sato, F, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-14 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the PsbP protein of photosystem II from Nicotiana tabacum
Embo Rep., 5, 2004
|
|
1V2Z
 
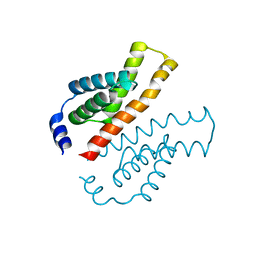 | | Crystal structure of the C-terminal domain of Thermosynechococcus elongatus BP-1 KaiA | | Descriptor: | circadian clock protein KaiA homolog | | Authors: | Uzumaki, T, Fujita, M, Nakatsu, T, Hayashi, F, Shibata, H, Itoh, N, Kato, H, Ishiura, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-20 | | Release date: | 2004-06-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the C-terminal clock-oscillator domain of the cyanobacterial KaiA protein
NAT.STRUCT.MOL.BIOL., 11, 2004
|
|
3WMG
 
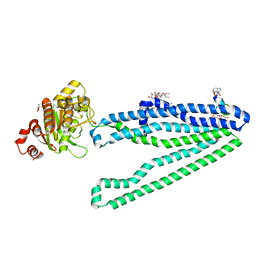 | | Crystal structure of an inward-facing eukaryotic ABC multidrug transporter G277V/A278V/A279V mutant in complex with an cyclic peptide inhibitor, aCAP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ATP-binding cassette, sub-family B, ... | | Authors: | Kodan, A, Yamaguchi, T, Nakatsu, T, Sakiyama, K, Hipolito, C.J, Fujioka, A, Hirokane, R, Ikeguchi, K, Watanabe, B, Hirtake, J, Kimura, Y, Suga, H, Ueda, K, Kato, H. | | Deposit date: | 2013-11-18 | | Release date: | 2014-04-30 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for gating mechanisms of a eukaryotic P-glycoprotein homolog.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WMF
 
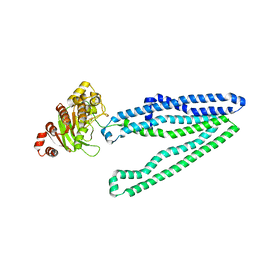 | | Crystal structure of an inward-facing eukaryotic ABC multitrug transporter G277V/A278V/A279V mutant | | Descriptor: | ATP-binding cassette, sub-family B, member 1, ... | | Authors: | Kodan, A, Yamaguchi, T, Nakatsu, T, Kato, H. | | Deposit date: | 2013-11-18 | | Release date: | 2014-03-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for gating mechanisms of a eukaryotic P-glycoprotein homolog.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WME
 
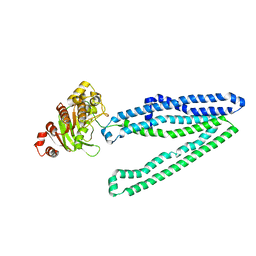 | | Crystal structure of an inward-facing eukaryotic ABC multidrug transporter | | Descriptor: | ATP-binding cassette, sub-family B, member 1, ... | | Authors: | Kodan, A, Yamaguchi, T, Nakatsu, T, Kato, H. | | Deposit date: | 2013-11-18 | | Release date: | 2014-03-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | Structural basis for gating mechanisms of a eukaryotic P-glycoprotein homolog.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2ZZO
 
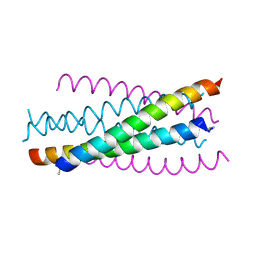 | | Crystal structure of the complex between GP41 fragment N36 and fusion inhibitor C34/S138A | | Descriptor: | Transmembrane protein | | Authors: | Watabe, T, Nakano, H, Nakatsu, T, Kato, H, Fujii, N. | | Deposit date: | 2009-02-20 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystallographic study of an HIV-1 fusion inhibitor with the gp41 S138A substitution
J.Mol.Biol., 392, 2009
|
|
3AJB
 
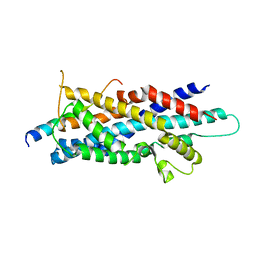 | | Crystal Structure of human Pex3p in complex with N-terminal Pex19p peptide | | Descriptor: | Peroxisomal biogenesis factor 19, Peroxisomal biogenesis factor 3 | | Authors: | Sato, Y, Shibata, H, Nakatsu, T, Kato, H. | | Deposit date: | 2010-05-27 | | Release date: | 2010-12-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for docking of peroxisomal membrane protein carrier Pex19p onto its receptor Pex3p
Embo J., 29, 2010
|
|
1IWQ
 
 | | Crystal Structure of MARCKS calmodulin binding domain peptide complexed with Ca2+/Calmodulin | | Descriptor: | CALCIUM ION, CALMODULIN, MARCKS | | Authors: | Yamauchi, E, Nakatsu, T, Matsubara, M, Kato, H, Taniguchi, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-05-31 | | Release date: | 2003-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a MARCKS peptide containing the calmodulin-binding domain in complex with Ca(2+)-calmodulin
NAT.STRUCT.BIOL., 10, 2003
|
|
1L7Z
 
 | | Crystal structure of Ca2+/Calmodulin complexed with myristoylated CAP-23/NAP-22 peptide | | Descriptor: | CALCIUM ION, CALMODULIN, CAP-23/NAP-22, ... | | Authors: | Matsubara, M, Nakatsu, T, Yamauchi, E, Kato, H, Taniguchi, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-18 | | Release date: | 2003-09-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a myristoylated CAP-23/NAP-22 N-terminal domain complexed with Ca2+/calmodulin
EMBO J., 23, 2004
|
|
3W15
 
 | | Structure of peroxisomal targeting signal 2 (PTS2) of Saccharomyces cerevisiae 3-ketoacyl-CoA thiolase in complex with Pex7p and Pex21p | | Descriptor: | 3-ketoacyl-CoA thiolase, peroxisomal, Maltose-binding periplasmic protein, ... | | Authors: | Pan, D, Nakatsu, T, Kato, H. | | Deposit date: | 2012-11-06 | | Release date: | 2013-07-03 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of peroxisomal targeting signal-2 bound to its receptor complex Pex7p-Pex21p
Nat.Struct.Mol.Biol., 20, 2013
|
|
