2G0D
 
 | | Nisin cyclase | | Descriptor: | Nisin biosynthesis protein nisC, ZINC ION | | Authors: | Nair, S.K. | | Deposit date: | 2006-02-12 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure and mechanism of the lantibiotic cyclase involved in nisin biosynthesis
Science, 311, 2006
|
|
2G02
 
 | | Nisin cyclase | | Descriptor: | Nisin biosynthesis protein nisC, ZINC ION | | Authors: | Nair, S.K. | | Deposit date: | 2006-02-10 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Mechanism of the Lantibiotic Cyclase Involved in Nisin Biosynthesis
Science, 311, 2006
|
|
7CA2
 
 | |
4H6V
 
 | |
12CA
 
 | |
1CA3
 
 | |
6WQ1
 
 | | Eukaryotic LanCL2 protein | | Descriptor: | LanC-like protein 2, ZINC ION | | Authors: | Nair, S.K, Garg, N. | | Deposit date: | 2020-04-28 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | LanCLs add glutathione to dehydroamino acids generated at phosphorylated sites in the proteome.
Cell, 184, 2021
|
|
6UAK
 
 | | LahSb - C-terminal methyltransferase involved in RiPP biosynthesis | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SAM dependent methyltransferase LahSB | | Authors: | Nair, S.K, Estrada, P. | | Deposit date: | 2019-09-10 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Characterization of a Dehydratase and Methyltransferase in the Biosynthesis of Ribosomally Synthesized and Post-translationally Modified Peptides in Lachnospiraceae.
Chembiochem, 21, 2020
|
|
6PGN
 
 | | PagF single mutant with GPP | | Descriptor: | GERANYL DIPHOSPHATE, MAGNESIUM ION, PagF | | Authors: | Nair, S.K, Hao, Y, Estrada, P. | | Deposit date: | 2019-06-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Single Amino Acid Switch Alters the Isoprene Donor Specificity in
Ribosomally Synthesized and Post-Translationally Modified Peptide
Prenyltransferases
J. Am. Chem. Soc., 140, 2018
|
|
6PGM
 
 | | PirF geranyltransferase | | Descriptor: | PirF Geranyltransferase | | Authors: | Nair, S.K, Hao, Y, Estrada, P. | | Deposit date: | 2019-06-24 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Single Amino Acid Switch Alters the Isoprene Donor Specificity in Ribosomally Synthesized and Post-Translationally Modified Peptide Prenyltransferases.
J.Am.Chem.Soc., 140, 2018
|
|
1HEB
 
 | |
1HEA
 
 | |
1HEC
 
 | |
1HCA
 
 | |
1HED
 
 | |
1OKL
 
 | | CARBONIC ANHYDRASE II COMPLEX WITH THE 1OKL INHIBITOR 5-DIMETHYLAMINO-NAPHTHALENE-1-SULFONAMIDE | | Descriptor: | 5-(DIMETHYLAMINO)-1-NAPHTHALENESULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Nair, S.K, Elbaum, D, Christianson, D.W. | | Deposit date: | 1996-06-25 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unexpected binding mode of the sulfonamide fluorophore 5-dimethylamino-1-naphthalene sulfonamide to human carbonic anhydrase II. Implications for the development of a zinc biosensor.
J.Biol.Chem., 271, 1996
|
|
8EJY
 
 | | [4+2] Aza-Cyclase F293A variant | | Descriptor: | PbtD | | Authors: | Nair, S.K. | | Deposit date: | 2022-09-19 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Enzymatic Pyridine Aromatization during Thiopeptide Biosynthesis.
J.Am.Chem.Soc., 144, 2022
|
|
8EJZ
 
 | | [4+2] Aza-Cyclase Y293F variant | | Descriptor: | PbtD | | Authors: | Nair, S.K. | | Deposit date: | 2022-09-19 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Enzymatic Pyridine Aromatization during Thiopeptide Biosynthesis.
J.Am.Chem.Soc., 144, 2022
|
|
1NLW
 
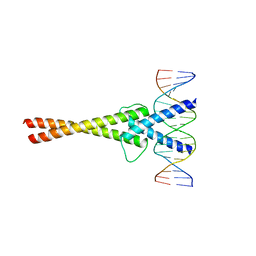 | | Crystal structure of Mad-Max recognizing DNA | | Descriptor: | 5'-D(*GP*AP*GP*TP*AP*GP*CP*AP*CP*GP*TP*GP*CP*TP*AP*CP*TP*C)-3', MAD PROTEIN, MAX PROTEIN | | Authors: | Nair, S.K, Burley, S.K. | | Deposit date: | 2003-01-07 | | Release date: | 2003-02-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of Myc-Max and Mad-Max recognizing DNA: Molecular bases of regulation by proto-oncogenic transcription factors
Cell(Cambridge,Mass.), 112, 2003
|
|
1YDC
 
 | |
1YDD
 
 | | STRUCTURAL BASIS OF INHIBITOR AFFINITY TO VARIANTS OF HUMAN CARBONIC ANHYDRASE II | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Nair, S.K, Christianson, D.W. | | Deposit date: | 1994-12-22 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of inhibitor affinity to variants of human carbonic anhydrase II.
Biochemistry, 34, 1995
|
|
1YDB
 
 | | STRUCTURAL BASIS OF INHIBITOR AFFINITY TO VARIANTS OF HUMAN CARBONIC ANHYDRASE II | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Nair, S.K, Christianson, D.W. | | Deposit date: | 1994-12-22 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of inhibitor affinity to variants of human carbonic anhydrase II.
Biochemistry, 34, 1995
|
|
4NU5
 
 | | Crystal Structure of PTDH R301A | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Phosphonate dehydrogenase | | Authors: | Nair, S.K, Chekan, J.R. | | Deposit date: | 2013-12-03 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chemical rescue and inhibition studies to determine the role of arg301 in phosphite dehydrogenase.
Plos One, 9, 2014
|
|
1YDA
 
 | | STRUCTURAL BASIS OF INHIBITOR AFFINITY TO VARIANTS OF HUMAN CARBONIC ANHYDRASE II | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Nair, S.K, Christianson, D.W. | | Deposit date: | 1994-12-22 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of inhibitor affinity to variants of human carbonic anhydrase II.
Biochemistry, 34, 1995
|
|
4OK4
 
 | | Crystal Structure of Alg17c Mutant H202L | | Descriptor: | Putative alginate lyase, ZINC ION | | Authors: | Nair, S.K, Park, D.S. | | Deposit date: | 2014-01-21 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a PL17 Family Alginate Lyase Demonstrates Functional Similarities among Exotype Depolymerases.
J.Biol.Chem., 289, 2014
|
|
