1K1Z
 
 | | Solution structure of N-terminal SH3 domain mutant(P33G) of murine Vav | | Descriptor: | vav | | Authors: | Ogura, K, Nagata, K, Horiuchi, M, Ebisui, E, Hasuda, T, Yuzawa, S, Nishida, M, Hatanaka, H, Inagaki, F. | | Deposit date: | 2001-09-26 | | Release date: | 2001-10-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of N-terminal SH3 domain of Vav and the recognition site for Grb2 C-terminal SH3 domain
J.BIOMOL.NMR, 22, 2002
|
|
1IX5
 
 | | Solution structure of the Methanococcus thermolithotrophicus FKBP | | Descriptor: | FKBP | | Authors: | Suzuki, R, Nagata, K, Kawakami, M, Nemoto, N, Furutani, M, Adachi, K, Maruyama, T, Tanokura, M. | | Deposit date: | 2002-06-12 | | Release date: | 2003-06-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional Solution Structure of an Archaeal FKBP with a Dual Function of Peptidyl Prolyl cis-trans Isomerase and Chaperone-like Activities
J.MOL.BIOL., 328, 2003
|
|
1GCQ
 
 | | CRYSTAL STRUCTURE OF VAV AND GRB2 SH3 DOMAINS | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2, VAV PROTO-ONCOGENE | | Authors: | Nishida, M, Nagata, K, Hachimori, Y, Ogura, K, Inagaki, F. | | Deposit date: | 2000-08-08 | | Release date: | 2001-08-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Novel recognition mode between Vav and Grb2 SH3 domains.
EMBO J., 20, 2001
|
|
1GCP
 
 | | CRYSTAL STRUCTURE OF VAV SH3 DOMAIN | | Descriptor: | VAV PROTO-ONCOGENE | | Authors: | Nishida, M, Nagata, K, Hachimori, Y, Ogura, K, Inagaki, F. | | Deposit date: | 2000-08-08 | | Release date: | 2001-08-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel recognition mode between Vav and Grb2 SH3 domains.
EMBO J., 20, 2001
|
|
2RQY
 
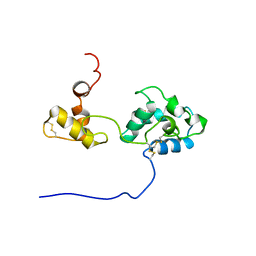 | | Solution structure and dynamics of mouse ARMET | | Descriptor: | Putative uncharacterized protein | | Authors: | Hoseki, J, Sasakawa, H, Yamaguchi, Y, Maeda, M, Kubota, H, Kato, K, Nagata, K. | | Deposit date: | 2010-01-26 | | Release date: | 2010-04-21 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of mouse ARMET.
Febs Lett., 584, 2010
|
|
4H8N
 
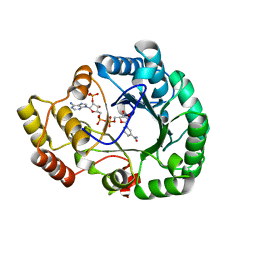 | | Crystal structure of conjugated polyketone reductase C2 from candida parapsilosis complexed with NADPH | | Descriptor: | Conjugated polyketone reductase C2, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Qin, H.-M, Yamamura, A, Miyakawa, T, Maruoka, S, Ohtsuka, J, Nagata, K, Kataoka, M, Shimizu, S, Tanokura, M. | | Deposit date: | 2012-09-23 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of conjugated polyketone reductase from Candida parapsilosis IFO 0708 reveals conformational changes for substrate recognition upon NADPH binding
Appl.Microbiol.Biotechnol., 98, 2014
|
|
1X25
 
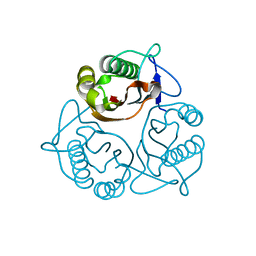 | | Crystal Structure of a Member of YjgF Family from Sulfolobus Tokodaii (ST0811) | | Descriptor: | Hypothetical UPF0076 protein ST0811 | | Authors: | Miyakawa, T, Lee, W.C, Hatano, K, Kato, Y, Sawano, Y, Miyazono, K, Nagata, K, Tanokura, M. | | Deposit date: | 2005-04-20 | | Release date: | 2006-02-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the YjgF/YER057c/UK114 family protein from the hyperthermophilic archaeon Sulfolobus tokodaii strain 7
Proteins, 62, 2006
|
|
1J0T
 
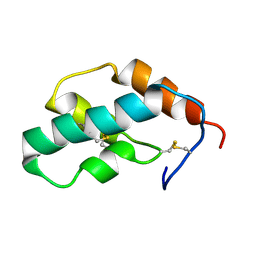 | | The solution structure of molt-inhibiting hormone from the kuruma prawn | | Descriptor: | MOLT-INHIBITING HORMONE | | Authors: | Katayama, H, Nagata, K, Ohira, T, Yumoto, F, Tanokura, M, Nagasawa, H. | | Deposit date: | 2002-11-22 | | Release date: | 2002-12-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of molt-inhibiting hormone from the Kuruma prawn Marsupenaeus japonicus
J.Biol.Chem., 278, 2003
|
|
3D79
 
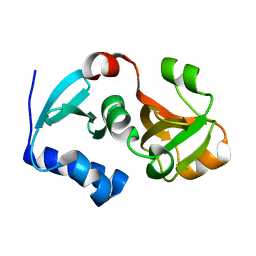 | | Crystal structure of hypothetical protein PH0734.1 from hyperthermophilic archaea Pyrococcus horikoshii OT3 | | Descriptor: | Putative uncharacterized protein PH0734 | | Authors: | Nishimura, Y, Miyazono, K, Sawano, Y, Makino, T, Nagata, K, Tanokura, M. | | Deposit date: | 2008-05-20 | | Release date: | 2008-12-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of hypothetical protein PH0734.1 from hyperthermophilic archaea Pyrococcus horikoshii OT3.
Proteins, 73, 2008
|
|
2DVY
 
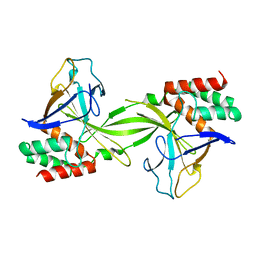 | | Crystal structure of restriction endonucleases PabI | | Descriptor: | Restriction endonuclease PabI | | Authors: | Miyazono, K, Watanabe, M, Kamo, M, Sawasaki, T, Nagata, K, Endo, Y, Tanokura, M, Kobayashi, I. | | Deposit date: | 2006-08-01 | | Release date: | 2007-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Novel protein fold discovered in the PabI family of restriction enzymes
Nucleic Acids Res., 35, 2007
|
|
3AI1
 
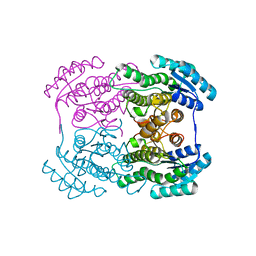 | | The crystal structure of L-sorbose reductase from Gluconobacter frateurii complexed with NADPH and L-sorbose reveals the structure bases of its catalytic mechanism and high substrate selectivity | | Descriptor: | NADPH-sorbose reductase | | Authors: | Kubota, K, Nagata, K, Okai, M, Miyazono, K, Tanokura, M. | | Deposit date: | 2010-05-06 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | The Crystal Structure of l-Sorbose Reductase from Gluconobacter frateurii Complexed with NADPH and l-Sorbose
J.Mol.Biol., 407, 2011
|
|
3AI3
 
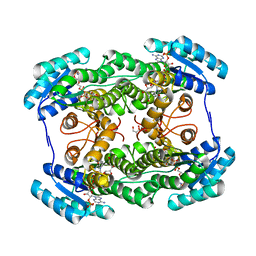 | | The crystal structure of L-Sorbose reductase from Gluconobacter frateurii complexed with NADPH and L-sorbose | | Descriptor: | L-sorbose, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-sorbose reductase, ... | | Authors: | Kubota, K, Nagata, K, Okai, M, Miyazono, K, Tanokura, M. | | Deposit date: | 2010-05-07 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of l-Sorbose Reductase from Gluconobacter frateurii Complexed with NADPH and l-Sorbose
J.Mol.Biol., 407, 2011
|
|
3AI2
 
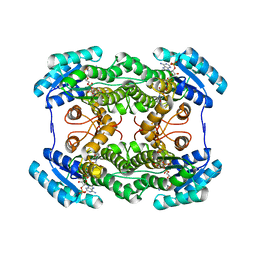 | | The crystal structure of L-sorbose reductase from Gluconobacter frateurii complexed with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-sorbose reductase | | Authors: | Kubota, K, Nagata, K, Okai, M, Miyazono, K, Tanokura, M. | | Deposit date: | 2010-05-07 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of l-Sorbose Reductase from Gluconobacter frateurii Complexed with NADPH and l-Sorbose
J.Mol.Biol., 407, 2011
|
|
3A03
 
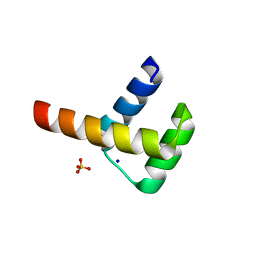 | | Crystal structure of Hox11L1 homeodomain | | Descriptor: | SODIUM ION, SULFATE ION, T-cell leukemia homeobox protein 2 | | Authors: | Miyazono, K, Nagata, K, Saigo, K, Kojima, T, Tanokura, M. | | Deposit date: | 2009-02-28 | | Release date: | 2010-03-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Cooperative DNA-binding and sequence-recognition mechanism of aristaless and clawless
Embo J., 29, 2010
|
|
3A01
 
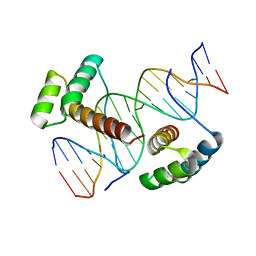 | | Crystal structure of Aristaless and Clawless homeodomains bound to DNA | | Descriptor: | 5'-D(*CP*CP*GP*CP*AP*AP*TP*TP*AP*AP*TP*TP*AP*AP*GP*CP*C)-3', 5'-D(*GP*GP*CP*TP*TP*AP*AP*TP*TP*AP*AP*TP*TP*GP*CP*GP*G)-3', Homeobox protein aristaless, ... | | Authors: | Miyazono, K, Nagata, K, Saigo, K, Kojima, T, Tanokura, M. | | Deposit date: | 2009-02-28 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cooperative DNA-binding and sequence-recognition mechanism of aristaless and clawless
Embo J., 29, 2010
|
|
3A02
 
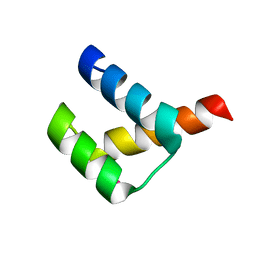 | | Crystal structure of Aristaless homeodomain | | Descriptor: | CADMIUM ION, CHLORIDE ION, Homeobox protein aristaless | | Authors: | Miyazono, K, Nagata, K, Saigo, K, Kojima, T, Tanokura, M. | | Deposit date: | 2009-02-28 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Cooperative DNA-binding and sequence-recognition mechanism of aristaless and clawless
Embo J., 29, 2010
|
|
2EGD
 
 | | Crystal structure of human S100A13 in the Ca2+-bound state | | Descriptor: | CALCIUM ION, Protein S100-A13 | | Authors: | Imai, F.L, Nagata, K, Yonezawa, N, Nakano, M, Tanokura, M. | | Deposit date: | 2007-02-28 | | Release date: | 2008-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human S100A13 in the Ca2+-bound state
Acta Crystallogr.,Sect.F, 64, 2008
|
|
3W6V
 
 | | Crystal structure of the DNA-binding domain of AdpA, the global transcriptional factor, in complex with a target DNA | | Descriptor: | AdpA, DNA (5'-D(*AP*GP*GP*TP*TP*GP*GP*CP*GP*GP*GP*TP*TP*CP*AP*C)-3'), DNA (5'-D(*CP*TP*GP*TP*GP*AP*AP*CP*CP*CP*GP*CP*CP*AP*AP*C)-3') | | Authors: | Yao, M.D, Ohtsuka, J, Nagata, K, Miyazono, K, Ohnishi, Y, Tanokura, M. | | Deposit date: | 2013-02-22 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Complex Structure of the DNA-binding Domain of AdpA, the Global Transcription Factor in Streptomyces griseus, and a Target Duplex DNA Reveals the Structural Basis of Its Tolerant DNA Sequence Specificity
J.Biol.Chem., 288, 2013
|
|
3WGB
 
 | | Crystal structure of aeromonas jandaei L-allo-threonine aldolase | | Descriptor: | GLYCINE, L-allo-threonine aldolase, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE] | | Authors: | Qin, H.M, Imai, F.L, Miyakawa, T, Kataoka, M, Okai, M, Ohtsuka, J, Hou, F, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2013-08-03 | | Release date: | 2014-07-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | L-allo-Threonine aldolase with an H128Y/S292R mutation from Aeromonas jandaei DK-39 reveals the structural basis of changes in substrate stereoselectivity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3APQ
 
 | |
3WGC
 
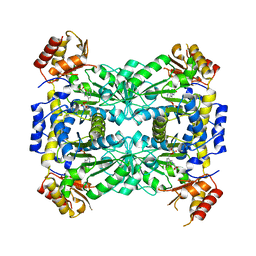 | | Aeromonas jandaei L-allo-threonine aldolase H128Y/S292R double mutant | | Descriptor: | L-allo-threonine aldolase, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE] | | Authors: | Qin, H.M, Imai, F.L, Miyakawa, T, Kataoka, M, Okai, M, Ohtsuka, J, Hou, F, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2013-08-03 | | Release date: | 2014-07-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | L-allo-Threonine aldolase with an H128Y/S292R mutation from Aeromonas jandaei DK-39 reveals the structural basis of changes in substrate stereoselectivity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3APO
 
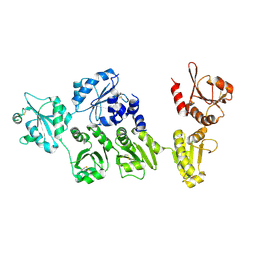 | | Crystal structure of full-length ERdj5 | | Descriptor: | DnaJ homolog subfamily C member 10 | | Authors: | Inaba, K, Suzuki, M, Nagata, K. | | Deposit date: | 2010-10-20 | | Release date: | 2011-04-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of an ERAD pathway mediated by the ER-resident protein disulfide reductase ERdj5.
Mol.Cell, 41, 2011
|
|
3APS
 
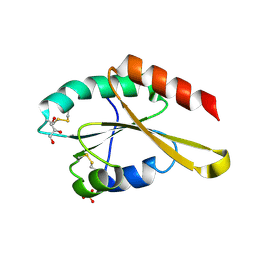 | | Crystal structure of Trx4 domain of ERdj5 | | Descriptor: | DnaJ homolog subfamily C member 10, GLYCEROL, SULFATE ION | | Authors: | Inaba, K, Suzuki, M, Nagata, K. | | Deposit date: | 2010-10-20 | | Release date: | 2011-04-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of an ERAD pathway mediated by the ER-resident protein disulfide reductase ERdj5.
Mol.Cell, 41, 2011
|
|
2EB1
 
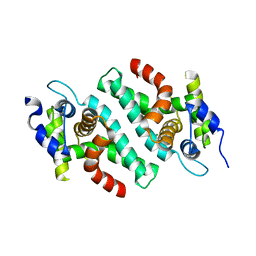 | | Crystal Structure of the C-Terminal RNase III Domain of Human Dicer | | Descriptor: | Endoribonuclease Dicer, MAGNESIUM ION | | Authors: | Takeshita, D, Zenno, S, Lee, W.C, Nagata, K, Saigo, K, Tanokura, M. | | Deposit date: | 2007-02-05 | | Release date: | 2007-11-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Homodimeric Structure and Double-stranded RNA Cleavage Activity of the C-terminal RNase III Domain of Human Dicer
J.Mol.Biol., 374, 2007
|
|
2Z1N
 
 | | Crystal structure of APE0912 from Aeropyrum pernix K1 | | Descriptor: | SODIUM ION, dehydrogenase | | Authors: | Ichimura, T, Yamamura, A, Mimoto, F, Ohtsuka, J, Miyazono, K, Okai, M, Kamo, M, Lee, W.-C, Nagata, K, Tanokura, M. | | Deposit date: | 2007-05-10 | | Release date: | 2008-03-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A unique catalytic triad revealed by the crystal structure of APE0912, a short-chain dehydrogenase/reductase family protein from Aeropyrum pernix K1
Proteins, 70, 2008
|
|
