6SWZ
 
 | |
8FTD
 
 | | Structure of Escherichia coli CedA in complex with transcription initiation complex | | Descriptor: | CHAPSO, Cell division activator CedA, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Liu, M, Vassyliev, N, Nudler, E. | | Deposit date: | 2023-01-11 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | General transcription factor from Escherichia coli with a distinct mechanism of action.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6RMW
 
 | | Structure of N-terminal truncated IMP bound Plasmodium falciparum IMP-nucleotidase | | Descriptor: | GLYCEROL, IMP-specific 5'-nucleotidase, putative, ... | | Authors: | Carrique, L, Ballut, L, Violot, S, Aghajari, N. | | Deposit date: | 2019-05-07 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure and catalytic regulation of Plasmodium falciparum IMP specific nucleotidase.
Nat Commun, 11, 2020
|
|
4B0S
 
 | | Structure of the Deamidase-Depupylase Dop of the Prokaryotic Ubiquitin-like Modification Pathway in Complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DEAMIDASE-DEPUPYLASE DOP, MAGNESIUM ION | | Authors: | Ozcelik, D, Barandun, J, Schmitz, N, Sutter, M, Guth, E, Damberger, F.F, Allain, F.H.-T, Ban, N, Weber-Ban, E. | | Deposit date: | 2012-07-04 | | Release date: | 2012-09-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structures of Pup Ligase Pafa and Depupylase Dop from the Prokaryotic Ubiquitin-Like Modification Pathway.
Nat.Commun., 3, 2012
|
|
6IMU
 
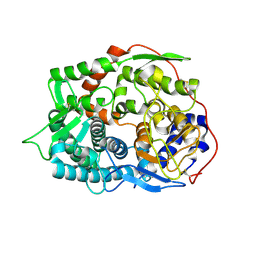 | | The apo-structure of endo-beta-1,2-glucanase from Talaromyces funiculosus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tanaka, N, Nakajima, M, Narukawa-Nara, M, Matsunaga, H, Kamisuki, S, Aramasa, H, Takahashi, Y, Sugimoto, N, Abe, K, Miyanaga, A, Yamashita, T, Sugawara, F, Kamakura, T, Komba, S, Nakai, H, Taguchi, H. | | Deposit date: | 2018-10-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification, characterization, and structural analyses of a fungal endo-beta-1,2-glucanase reveal a new glycoside hydrolase family.
J.Biol.Chem., 294, 2019
|
|
7CTS
 
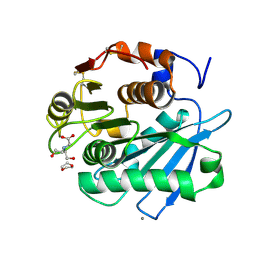 | | Open form of PET-degrading cutinase Cut190 with thermostability-improving mutations of S226P/R228S/Q138A/D250C-E296C/Q123H/N202H and S176A inactivation | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Alpha/beta hydrolase family protein, BICINE, ... | | Authors: | Emori, M, Numoto, N, Senga, A, Bekker, G.J, Kamiya, N, Ito, N, Kawai, F, Oda, M. | | Deposit date: | 2020-08-20 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis of mutants of PET-degrading enzyme from Saccharomonospora viridis AHK190 with high activity and thermal stability.
Proteins, 89, 2021
|
|
4CL7
 
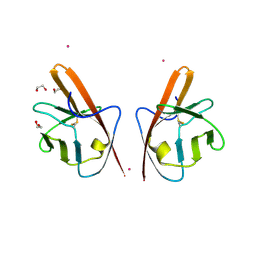 | | Crystal structure of VEGFR-1 domain 2 in presence of Cobalt | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR 1 | | Authors: | Gaucher, J.-F, Reille-Seroussi, M, Gagey-Eilstein, N, Broussy, S, Coric, P, Seijo, B, Lascombe, M.-B, Gautier, B, Liu, W.-Q, Huguenot, F, Inguimbert, N, Bouaziz, S, Vidal, M, Broutin, I. | | Deposit date: | 2014-01-13 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biophysical Studies of the Induced Dimerization of Human Vegf R Receptor 1 Binding Domain by Divalent Metals Competing with Vegf-A
Plos One, 11, 2016
|
|
2NBI
 
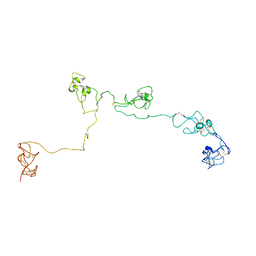 | | Structure of the PSCD-region of the cell wall protein pleuralin-1 | | Descriptor: | HEP200 protein | | Authors: | De Sanctis, S, Wenzler, M, Kroeger, N, Malloni, W.M, Sumper, M, Rainer, D, Zadravec, P, Brunner, E, Kremer, W, Kalbitzer, H.R. | | Deposit date: | 2016-02-23 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | PSCD Domains of Pleuralin-1 from the Diatom Cylindrotheca fusiformis: NMR Structures and Interactions with Other Biosilica-Associated Proteins.
Structure, 24, 2016
|
|
6SGZ
 
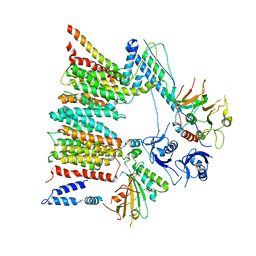 | | Structure of protomer 2 of the ESX-3 core complex | | Descriptor: | ESX-3 secretion system ATPase EccB3, ESX-3 secretion system protein EccC3, ESX-3 secretion system protein EccD3, ... | | Authors: | Famelis, N, Rivera-Calzada, A, Llorca, O, Geibel, S. | | Deposit date: | 2019-08-05 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Architecture of the mycobacterial type VII secretion system.
Nature, 576, 2019
|
|
6Z1A
 
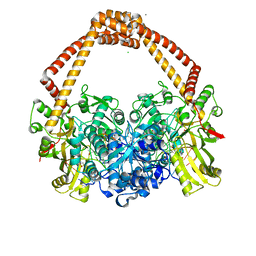 | | Ternary complex of Staphylococcus aureus DNA gyrase with AMK12 and DNA | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Kolaric, A, Germe, T, Hrast, M, Stevenson, C.E.M, Lawson, D.M, Burton, N, Voros, J, Maxwell, A, Minovski, N, Anderluh, M. | | Deposit date: | 2020-05-13 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Potent DNA gyrase inhibitors bind asymmetrically to their target using symmetrical bifurcated halogen bonds.
Nat Commun, 12, 2021
|
|
6JT7
 
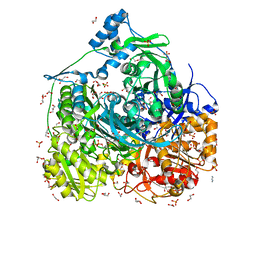 | | Crystal structure of 452-453_deletion mutant of FGAM Synthetase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Sharma, N, Ahalawat, N, Sandhu, P, Mondal, J, Anand, R. | | Deposit date: | 2019-04-10 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Role of allosteric switches and adaptor domains in long-distance cross-talk and transient tunnel formation.
Sci Adv, 6, 2020
|
|
6JSN
 
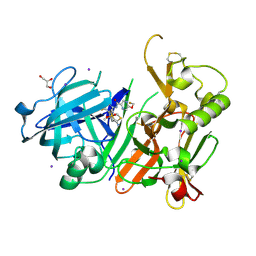 | | Crystal Structure of BACE1 in complex with N-{3-[(5R)-3-amino-5-methyl-9,9-dioxo-2,9lambda6-dithia-4-azaspiro[5.5]undec-3-en-5-yl]-4-fluorophenyl}-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Fujimoto, K, Matsuoka, E, Asada, N, Tadano, G, Yamamoto, T, Nakahara, K, Fuchino, K, Ito, H, Kanegawa, N, Moechars, D, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Design of Selective beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors: Targeting the Flap to Gain Selectivity over BACE2.
J.Med.Chem., 62, 2019
|
|
4CQE
 
 | | B-Raf Kinase V600E mutant in complex with a diarylthiazole B-Raf Inhibitor | | Descriptor: | N-{4-[2-(1-cyclopropylpiperidin-4-yl)-4-(3-{[(2,5-difluorophenyl)sulfonyl]amino}-2-fluorophenyl)-1,3-thiazol-5-yl]pyridin-2-yl}acetamide, SLC45A3-BRAF FUSION PROTEIN | | Authors: | Casale, E, Fasolini, M, Pulici, M, Traquandi, G, Marchionni, C, Modugno, M, Lupi, R, Amboldi, N, Colombo, N, Corti, L, Gasparri, F, Pastori, W, Scolaro, A, Donati, D, Felder, E, Galvani, A, Isacchi, A, Pesenti, E, Ciomei, M. | | Deposit date: | 2014-02-14 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Optimization of Diarylthiazole B-Raf Inhibitors: Identification of a Compound Endowed with High Oral Antitumor Activity, Mitigated Herg Inhibition, and Low Paradoxical Effect.
Chemmedchem, 10, 2015
|
|
6TKX
 
 | | Carbohydrate esterase from gut microbiota | | Descriptor: | Carbohydrate esterase, SULFATE ION | | Authors: | Penttinen, L, Hakulinen, N, Master, R.E. | | Deposit date: | 2019-11-29 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Polysaccharide utilization loci-driven enzyme discovery reveals BD-FAE: a bifunctional feruloyl and acetyl xylan esterase active on complex natural xylans.
Biotechnol Biofuels, 14, 2021
|
|
4CWP
 
 | | Human HSP90 alpha N-terminal domain in complex with an Aminotriazoloquinazoline inhibitor | | Descriptor: | 5-(1,3-benzodioxol-5-ylmethyl)[1,2,4]triazolo[1,5-c]quinazolin-2-amine, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Casale, E, Amboldi, N, Brasca, G, Caronni, D, Colombo, N, Dalvit, C, Felder, E.R, Fogliatto, G, Isacchi, A, Mantegani, S, Polucci, P, Riceputi, L, Sola, F, Visco, C, Zuccotto, F, Casuscelli, F. | | Deposit date: | 2014-04-03 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fragment-Based Hit Discovery and Structure-Based Optimization of Aminotriazoloquinazolines as Novel Hsp90 Inhibitors.
Bioorg.Med.Chem., 22, 2014
|
|
6J34
 
 | | Crystal Structure of maltotriose-complex of PulA from Klebsiella pneumoniae | | Descriptor: | ACETATE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2019-01-09 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Relationship between the induced-fit loop and the activity of Klebsiella pneumoniae pullulanase.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6JSE
 
 | | Crystal Structure of BACE1 in complex with N-(3-((4S,5R)-2-amino-4-methyl-5-phenyl-5,6-dihydro-4H-1,3-thiazin-4-yl)-4-fluorophenyl)-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, IODIDE ION, ... | | Authors: | Fujimoto, K, Matsuoka, E, Asada, N, Tadano, G, Yamamoto, T, Nakahara, K, Fuchino, K, Ito, H, Kanegawa, N, Moechars, D, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design of Selective beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors: Targeting the Flap to Gain Selectivity over BACE2.
J.Med.Chem., 62, 2019
|
|
6KW3
 
 | | The ClassA RSC-Nucleosome Complex | | Descriptor: | Actin-like protein ARP9, Actin-related protein 7, Chromatin structure-remodeling complex protein RSC3, ... | | Authors: | Ye, Y.P, Wu, H, Chen, K.J, Verma, N, Cairns, B, Gao, N, Chen, Z.C. | | Deposit date: | 2019-09-05 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.13 Å) | | Cite: | Structure of the RSC complex bound to the nucleosome.
Science, 366, 2019
|
|
4BQG
 
 | | structure of HSP90 with an inhibitor bound | | Descriptor: | 5-(3,4-dichloro-phenoxy)-benzene-1,3-diol, HSP90AA1 PROTEIN | | Authors: | Casale, E, Brasca, M.G, Mantegani, S, Amboldi, N, Bindi, S, Caronni, D, Ceccarelli, W, Colombo, N, DePonti, A, Donati, D, Ermoli, A, Fachin, G, Felder, E.R, Ferguson, R.D, Fiorelli, C, Guanci, M, Isacchi, A, Pesenti, E, Polucci, P, Riceputi, L, Sola, F, Visco, C, Zuccotto, F, Fogliatto, G. | | Deposit date: | 2013-05-30 | | Release date: | 2013-10-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Nms-E973 as Novel, Selective and Potent Inhibitor of Heat Shock Protein 90 (Hsp90).
Bioorg.Med.Chem., 21, 2013
|
|
1AZE
 
 | | NMR STRUCTURE OF THE COMPLEX BETWEEN THE C32S-Y7V MUTANT OF THE NSH3 DOMAIN OF GRB2 WITH A PEPTIDE FROM SOS, 10 STRUCTURES | | Descriptor: | GRB2, SOS | | Authors: | Vidal, M, Gincel, E, Goudreau, N, Cornille, F, Parker, F, Duchesne, M, Tocque, B, Garbay, C, Roques, B.P. | | Deposit date: | 1997-11-17 | | Release date: | 1999-05-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular and cellular analysis of Grb2 SH3 domain mutants: interaction with Sos and dynamin.
J.Mol.Biol., 290, 1999
|
|
1B0I
 
 | | ALPHA-AMYLASE FROM ALTEROMONAS HALOPLANCTIS | | Descriptor: | CALCIUM ION, CHLORIDE ION, PROTEIN (ALPHA-AMYLASE) | | Authors: | Aghajari, N, Haser, R. | | Deposit date: | 1998-11-10 | | Release date: | 1999-11-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the psychrophilic Alteromonas haloplanctis alpha-amylase give insights into cold adaptation at a molecular level.
Structure, 6, 1998
|
|
6KR6
 
 | | Crystal structure of Drosophila Piwi | | Descriptor: | MERCURY (II) ION, Protein piwi, ZINC ION, ... | | Authors: | Yamaguchi, S, Oe, A, Yamashita, K, Hirano, S, Mastumoto, N, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2019-08-21 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of Drosophila Piwi.
Nat Commun, 11, 2020
|
|
7CTR
 
 | | Closed form of PET-degrading cutinase Cut190 with thermostability-improving mutations of S226P/R228S/Q138A/D250C-E296C/Q123H/N202H | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Alpha/beta hydrolase family protein | | Authors: | Emori, M, Numoto, N, Senga, A, Bekker, G.J, Kamiya, N, Ito, N, Kawai, F, Oda, M. | | Deposit date: | 2020-08-20 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis of mutants of PET-degrading enzyme from Saccharomonospora viridis AHK190 with high activity and thermal stability.
Proteins, 89, 2021
|
|
4CWS
 
 | | Human HSP90 alpha N-terminal domain in complex with an Aminotriazoloquinazoline inhibitor | | Descriptor: | 2-{[2-amino-5-(1,3-benzodioxol-5-ylmethyl)[1,2,4]triazolo[1,5-c]quinazolin-8-yl]amino}ethanol, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Casale, E, Amboldi, N, Brasca, G, Caronni, D, Colombo, N, Dalvit, C, Felder, E.R, Fogliatto, G, Isacchi, A, Mantegani, S, Polucci, P, Riceputi, L, Sola, F, Visco, C, Zuccotto, F, Casuscelli, F. | | Deposit date: | 2014-04-03 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fragment-Based Hit Discovery and Structure-Based Optimization of Aminotriazoloquinazolines as Novel Hsp90 Inhibitors.
Bioorg.Med.Chem., 22, 2014
|
|
4CWR
 
 | | Human HSP90 alpha N-terminal domain in complex with an Aminotriazoloquinazoline inhibitor | | Descriptor: | 5-(1,3-benzodioxol-5-ylmethyl)-10-fluoro[1,2,4]triazolo[1,5-c]quinazolin-2-amine, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Casale, E, Amboldi, N, Brasca, G, Caronni, D, Colombo, N, Dalvit, C, Felder, E.R, Fogliatto, G, Isacchi, A, Mantegani, S, Polucci, P, Riceputi, L, Sola, F, Visco, C, Zuccotto, F, Casuscelli, F. | | Deposit date: | 2014-04-03 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-Based Hit Discovery and Structure-Based Optimization of Aminotriazoloquinazolines as Novel Hsp90 Inhibitors.
Bioorg.Med.Chem., 22, 2014
|
|
