5CBV
 
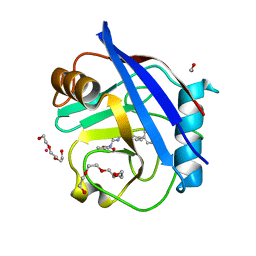 | | Human Cyclophilin D Complexed with Inhibitor | | Descriptor: | FORMIC ACID, Human Cyclophilin D, POTASSIUM ION, ... | | Authors: | Gibson, R.P, Shore, E, Kershaw, N, Awais, M, Javed, A, Latawiec, D, Pandalaneni, S, Wen, L, Berry, N, O'Neill, P, Sutton, R, Lian, L.Y. | | Deposit date: | 2015-07-01 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human Cyclophilin D Complexed with Inhibitor
To Be Published
|
|
1E6O
 
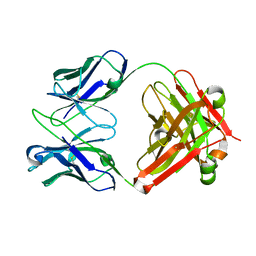 | | Crystal structure of Fab13B5 against HIV-1 capsid protein p24 | | Descriptor: | IMMUNOGLOBULIN HEAVY CHAIN, IMMUNOGLOBULIN LIGHT CHAIN | | Authors: | Monaco-Malbet, S, Berthet-Colominas, C, Novelli, A, Battai, N, Piga, N, Mallet, F, Cusack, S. | | Deposit date: | 2000-08-21 | | Release date: | 2000-11-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mutual Conformational Adaptations in Antigen and Antibody Upon Complex Formation between an Fab and HIV-1 Capsid Protein P24
Structure, 8, 2000
|
|
7NFV
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Srinivasan, V, Gunther, S, Reinke, P, Werner, N, Falke, S, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Sprenger, J, Lieske, J, Ginn, H, Lane, T.J, Yefanov, O, Gelisio, L, Koua, F, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Ewert, W, Trost, F, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Ehrt, C, Rarey, M, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Hinrichs, W, Meents, A, Betzel, C. | | Deposit date: | 2021-02-07 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
5CM2
 
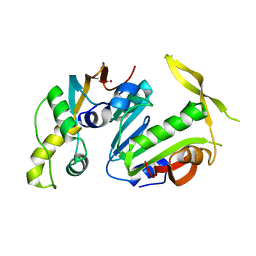 | | Structure of Y. lipolytica Trm9-Trm112 complex, a methyltransferase modifying U34 in the anticodon loop of some tRNAs | | Descriptor: | TRNA METHYLTRANSFERASE, TRNA METHYLTRANSFERASE ACTIVATOR SUBUNIT, ZINC ION | | Authors: | Letoquart, J, van Tran, N, Caroline, V, Aleksandrov, A, Lazar, N, Van Tilbeurgh, H, Liger, D, Graille, M. | | Deposit date: | 2015-07-16 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into molecular plasticity in protein complexes from Trm9-Trm112 tRNA modifying enzyme crystal structure.
Nucleic Acids Res., 43, 2015
|
|
7OVC
 
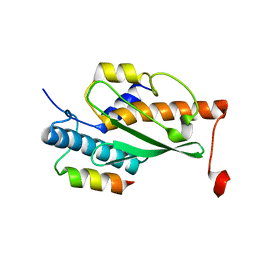 | | Structure of the human UFC1 protein in complex with the UBA5 C-terminal UFC1-binding motif. | | Descriptor: | Ubiquitin-fold modifier-conjugating enzyme 1, Ubiquitin-like modifier-activating enzyme 5 | | Authors: | Wesch, W, Loehr, F, Rogova, N, Doetsch, V, Rogov, V.V. | | Deposit date: | 2021-06-14 | | Release date: | 2021-08-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A Concerted Action of UBA5 C-Terminal Unstructured Regions Is Important for Transfer of Activated UFM1 to UFC1.
Int J Mol Sci, 22, 2021
|
|
5CBW
 
 | | Human Cyclophilin D Complexed with Inhibitor. | | Descriptor: | FORMIC ACID, POTASSIUM ION, Peptidyl-prolyl cis-trans isomerase F, ... | | Authors: | Gibson, R.P, Shore, E, Kershaw, N, Awais, M, Javed, A, Latawiec, D, Pandalaneni, S, Wen, L, Berry, N, O'Neill, P, Sutton, R, Lian, L.Y. | | Deposit date: | 2015-07-01 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human Cyclophilin D Complexed with Inhibitor.
To Be Published
|
|
2RDO
 
 | | 50S subunit with EF-G(GDPNP) and RRF bound | | Descriptor: | 23S RIBOSOMAL RNA, 50S ribosomal protein L1, 50S ribosomal protein L11, ... | | Authors: | Gao, N, Zavialov, A.V, Ehrenberg, M, Frank, J. | | Deposit date: | 2007-09-24 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Specific interaction between EF-G and RRF and its implication for GTP-dependent ribosome splitting into subunits.
J.Mol.Biol., 374, 2007
|
|
5CCQ
 
 | | Human Cyclophilin D Complexed with Inhibitor | | Descriptor: | FORMIC ACID, POTASSIUM ION, Peptidyl-prolyl cis-trans isomerase F, ... | | Authors: | Gibson, R.P, Shore, E, Kershaw, N, Awais, M, Javed, A, Latawiec, D, Pandalaneni, S, Wen, L, Berry, N, O'Neill, P, Sutton, R, Lian, L.Y. | | Deposit date: | 2015-07-02 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human Cyclophilin D Complexed with Inhibitor
To Be Published
|
|
1AIF
 
 | | ANTI-IDIOTYPIC FAB 409.5.3 (IGG2A) FAB FROM MOUSE | | Descriptor: | ANTI-IDIOTYPIC FAB 409.5.3 (IGG2A) FAB (HEAVY CHAIN), ANTI-IDIOTYPIC FAB 409.5.3 (IGG2A) FAB (LIGHT CHAIN) | | Authors: | Ban, N, Escobar, C, Hasel, K, Day, J, Greenwood, A, McPherson, A. | | Deposit date: | 1994-11-14 | | Release date: | 1997-02-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of an anti-idiotypic Fab against feline peritonitis virus-neutralizing antibody and a comparison with the complexed Fab.
FASEB J., 9, 1995
|
|
1ARX
 
 | | CRYSTAL STRUCTURES OF CYANIDE-AND TRIIODIDE-BOUND FORMS OF ARTHROMYCES RAMOSUS PEROXIDASE AT DIFFERENT PH VALUES. PERTURBATIONS OF ACTIVE SITE RESIDUES AND THEIR IMPLICATION IN ENZYME CATALYSIS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Fukuyama, K, Kunishima, N, Amada, F. | | Deposit date: | 1995-04-25 | | Release date: | 1996-01-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of cyanide- and triiodide-bound forms of Arthromyces ramosus peroxidase at different pH values. Perturbations of active site residues and their implication in enzyme catalysis.
J.Biol.Chem., 270, 1995
|
|
1ARY
 
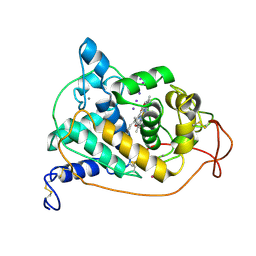 | | CRYSTAL STRUCTURES OF CYANIDE-AND TRIIODIDE-BOUND FORMS OF ARTHROMYCES RAMOSUS PEROXIDASE AT DIFFERENT PH VALUES. PERTURBATIONS OF ACTIVE SITE RESIDUES AND THEIR IMPLICATION IN ENZYME CATALYSIS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Fukuyama, K, Kunishima, N, Amada, F. | | Deposit date: | 1995-04-25 | | Release date: | 1996-01-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of cyanide- and triiodide-bound forms of Arthromyces ramosus peroxidase at different pH values. Perturbations of active site residues and their implication in enzyme catalysis.
J.Biol.Chem., 270, 1995
|
|
1AQH
 
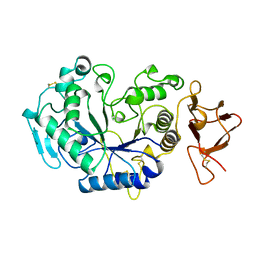 | | ALPHA-AMYLASE FROM ALTEROMONAS HALOPLANCTIS | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, CHLORIDE ION | | Authors: | Aghajari, N, Haser, R. | | Deposit date: | 1997-07-30 | | Release date: | 1999-02-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the psychrophilic alpha-amylase from Alteromonas haloplanctis in its native form and complexed with an inhibitor.
Protein Sci., 7, 1998
|
|
7Q1V
 
 | |
1ARP
 
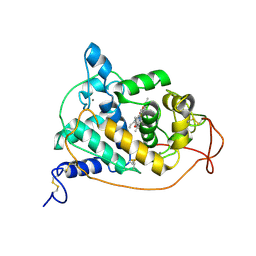 | |
6Q8J
 
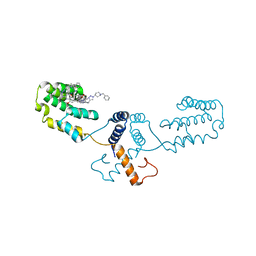 | | Nterminal domain of human SMU1 in complex with LSP641 | | Descriptor: | 1-~{tert}-butyl-3-[6-(3,5-dimethoxyphenyl)-2-[[1-[1-(phenylmethyl)piperidin-4-yl]-1,2,3-triazol-4-yl]methylamino]pyrido[2,3-d]pyrimidin-7-yl]urea, WD40 repeat-containing protein SMU1 | | Authors: | Tengo, L, Le Corre, L, Fournier, G, Ashraf, U, Busca, P, Rameix-Welti, M.-A, Gravier-Pelletier, C, Ruigrok, R.W.H, Jacob, Y, Vidalain, P.-O, Pietrancosta, N, Naffakh, N, McCarthy, A.A, Crepin, T. | | Deposit date: | 2018-12-14 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Destabilization of the human RED-SMU1 splicing complex as a basis for host-directed antiinfluenza strategy.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8BVA
 
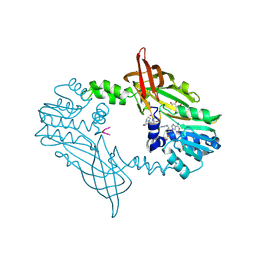 | | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 in complex with RSF1_114-126 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, L(+)-TARTARIC ACID, POTASSIUM ION, ... | | Authors: | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2022-12-02 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 in complex with RSF1_114-126
To Be Published
|
|
8BR6
 
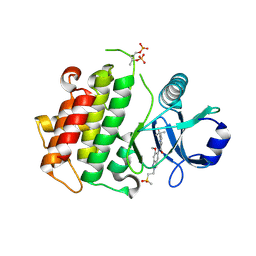 | | Discovery of IRAK4 Inhibitor 40 | | Descriptor: | ACETATE ION, Interleukin-1 receptor-associated kinase 4, ~{N}-[6-methoxy-2-(2-methylsulfonylethyl)-1,3-dihydroindazol-5-yl]-6-(2-oxidanylpropan-2-yl)pyridine-2-carboxamide | | Authors: | Schafer, M, Bothe, U, Schmidt, N, Gunther, J, Nubbemeyer, R, Siebeneicher, H, Ring, S, Boemer, U, Peters, M, Denner, K, Himmel, H, Sutter, A, Terebesi, I, Lange, M, Wengner, A.M, Guimond, N, Thaler, T, Platzek, J, Ewerspaecher, U, Steuber, H, Steinmeyer, A, Zollner, T.M. | | Deposit date: | 2022-11-22 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.167 Å) | | Cite: | Discovery of IRAK4 Inhibitors BAY1834845 (Zabedosertib) and BAY1830839 .
J.Med.Chem., 67, 2024
|
|
8BR5
 
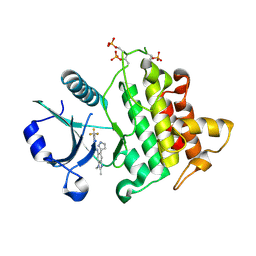 | | Discovery of IRAK4 Inhibitor 41 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, ~{N}-[2,3-dimethyl-6-(1~{H}-pyrazol-5-yl)benzimidazol-5-yl]-6-(trifluoromethyl)pyridine-2-carboxamide | | Authors: | Schafer, M, Bothe, U, Schmidt, N, Gunther, J, Nubbemeyer, R, Siebeneicher, H, Ring, S, Boemer, U, Peters, M, Denner, K, Himmel, H, Sutter, A, Terebesi, I, Lange, M, Wenger, A.M, Guimond, N, Thaler, T, Platzek, J, Eberspaecher, U, Steuber, H, Steinmeyer, A, Zollner, T.M. | | Deposit date: | 2022-11-22 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of IRAK4 Inhibitors BAY1834845 (Zabedosertib) and BAY1830839 .
J.Med.Chem., 67, 2024
|
|
8BR7
 
 | | Discovery of IRAK4 Inhibitors BAY1834845 and BAY1830839 | | Descriptor: | 3-nitro-~{N}-[2-[2-oxidanylidene-2-[4-(phenylcarbonyl)piperazin-1-yl]ethyl]indazol-5-yl]benzamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Schafer, M, Bothe, U, Schmidt, N, Gunther, J, Nubbemeyer, R, Siebeneicher, H, Ring, S, Boemer, U, Peters, M, Denner, K, Himmel, H, Sutter, A, Terebesi, I, Lange, M, Wenger, A.M, Guimond, N, Thaler, T, Platzek, J, Eberspaecher, U, Steuber, H, Steinmeyer, A, Zollner, T.M. | | Deposit date: | 2022-11-22 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.119 Å) | | Cite: | Discovery of IRAK4 Inhibitors BAY1834845 (Zabedosertib) and BAY1830839 .
J.Med.Chem., 67, 2024
|
|
6Q8I
 
 | | Nterminal domain of human SMU1 in complex with human REDmid | | Descriptor: | Protein Red, WD40 repeat-containing protein SMU1 | | Authors: | Tengo, L, Le Corre, L, Fournier, G, Ashraf, U, Busca, P, Rameix-Welti, M.-A, Gravier-Pelletier, C, Ruigrok, R.W.H, Jacob, Y, Vidalain, P.-O, Pietrancosta, N, Naffakh, N, McCarthy, A.A, Crepin, T. | | Deposit date: | 2018-12-14 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Destabilization of the human RED-SMU1 splicing complex as a basis for host-directed antiinfluenza strategy.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7PHZ
 
 | | Crystal structure of X77 bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in spacegroup P2(1)2(1)2(1). | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2021-08-19 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
8CAT
 
 | | The NADPH binding site on beef liver catalase | | Descriptor: | CATALASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Murthy, M.R.N, Reid III, T.J, Sicignano, A, Tanaka, N, Fita, I, Rossmann, M.G. | | Deposit date: | 1984-11-15 | | Release date: | 1985-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The NADPH binding site on beef liver catalase.
Proc.Natl.Acad.Sci.USA, 82, 1985
|
|
2RSM
 
 | | Solution structure and siRNA-mediated knockdown analysis of the mitochondrial disease-related protein C12orf65 (ICT2) | | Descriptor: | Probable peptide chain release factor C12orf65 homolog, mitochondrial | | Authors: | Enomoto, M, Tochio, N, Tomizawa, T, Koshiba, S, Guntert, P, Kigawa, T, Yokoyama, S, Nameki, N. | | Deposit date: | 2012-03-28 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure and siRNA-mediated knockdown analysis of the mitochondrial disease-related protein C12orf65.
Proteins, 2012
|
|
1FLM
 
 | | DIMER OF FMN-BINDING PROTEIN FROM DESULFOVIBRIO VULGARIS (MIYAZAKI F) | | Descriptor: | FLAVIN MONONUCLEOTIDE, PROTEIN (FMN-BINDING PROTEIN) | | Authors: | Suto, K, Kawagoe, K, Shibata, N, Morimoto, K, Higuchi, Y, Kitamura, M, Nakaya, T, Yasuoka, N. | | Deposit date: | 1999-03-10 | | Release date: | 2000-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | How do the x-ray structure and the NMR structure of FMN-binding protein differ?
Acta Crystallogr.,Sect.D, 56, 2000
|
|
2RPR
 
 | | Solution structure of the fifth FLYWCH domain of FLYWCH-type zinc finger-containing protein 1 | | Descriptor: | FLYWCH-type zinc finger-containing protein 1, ZINC ION | | Authors: | Enomoto, M, Saito, K, Tochio, N, Kigawa, T, Yokoyama, S, Nameki, N. | | Deposit date: | 2008-07-24 | | Release date: | 2009-08-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the fifth FLYWCH domain of FLYWCH-type zinc finger-containing protein 1
To be Published
|
|
