5H41
 
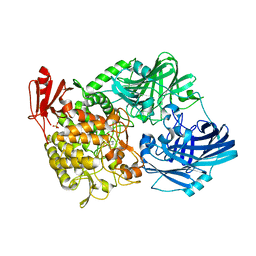 | | Crystal Structure of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans in complex with sophorose, isofagomine, sulfate ion | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, SULFATE ION, Uncharacterized protein, ... | | Authors: | Nakajima, M, Tanaka, N, Furukawa, N, Nihira, T, Kodutsumi, Y, Takahashi, Y, Sugimoto, N, Miyanaga, A, Fushinobu, S, Taguchi, H, Nakai, H. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic insight into the substrate specificity of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans
Sci Rep, 7, 2017
|
|
5GVQ
 
 | | Solution structure of the first RRM domain of human spliceosomal protein SF3b49 | | Descriptor: | Splicing factor 3B subunit 4 | | Authors: | Kuwasako, K, Nameki, N, Tsuda, K, Takahashi, M, Sato, A, Tochio, N, Inoue, M, Terada, T, Kigawa, T, Kobayashi, N, Shirouzu, M, Ito, T, Sakamoto, T, Wakamatsu, K, Guntert, P, Takahashi, S, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2016-09-06 | | Release date: | 2017-04-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first RNA recognition motif domain of human spliceosomal protein SF3b49 and its mode of interaction with a SF3b145 fragment.
Protein Sci., 26, 2017
|
|
3KDJ
 
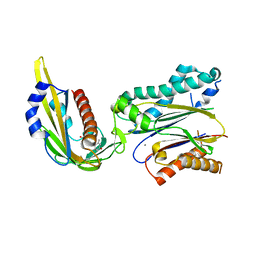 | | Complex structure of (+)-ABA-bound PYL1 and ABI1 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, MANGANESE (II) ION, Protein phosphatase 2C 56, ... | | Authors: | Yin, P, Fan, H, Hao, Q, Yuan, X, Yan, N. | | Deposit date: | 2009-10-23 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.878 Å) | | Cite: | Structural insights into the mechanism of abscisic acid signaling by PYL proteins
Nat.Struct.Mol.Biol., 16, 2009
|
|
3KD8
 
 | |
5H40
 
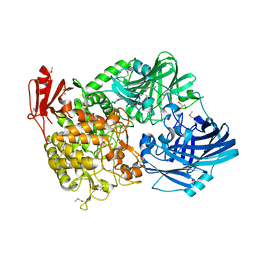 | | Crystal Structure of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans in complex with sophorose | | Descriptor: | CALCIUM ION, GLYCEROL, Uncharacterized protein, ... | | Authors: | Nakajima, M, Tanaka, N, Furukawa, N, Nihira, T, Kodutsumi, Y, Takahashi, Y, Sugimoto, N, Miyanaga, A, Fushinobu, S, Taguchi, H, Nakai, H. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic insight into the substrate specificity of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans
Sci Rep, 7, 2017
|
|
3KEX
 
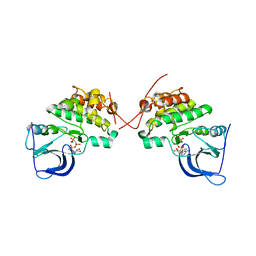 | | Crystal structure of the catalytically inactive kinase domain of the human epidermal growth factor receptor 3 (HER3) | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Receptor tyrosine-protein kinase erbB-3 | | Authors: | Jura, N, Shan, Y, Cao, X, Shaw, D.E, Kuriyan, J. | | Deposit date: | 2009-10-26 | | Release date: | 2009-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | Structural analysis of the catalytically inactive kinase domain of the human EGF receptor 3.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3KEG
 
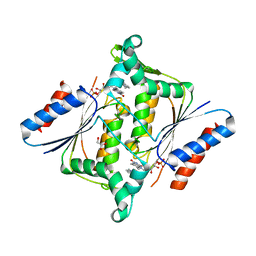 | | X-ray Crystallographic Structure of a Y131F mutant of Pseudomonas Aeruginosa Azoreductase in complex with Methyl RED | | Descriptor: | 2-(4-DIMETHYLAMINOPHENYL)DIAZENYLBENZOIC ACID, FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase 1, ... | | Authors: | Wang, C.-J, Laurieri, N, Abuhammad, A, Lowe, E, Westwood, I, Ryan, A, Sim, E. | | Deposit date: | 2009-10-26 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Role of tyrosine 131 in the active site of paAzoR1, an azoreductase with specificity for the inflammatory bowel disease prodrug balsalazide
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3KMP
 
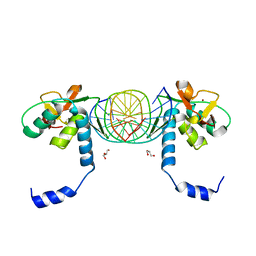 | | Crystal Structure of SMAD1-MH1/DNA complex | | Descriptor: | 5'-D(P*AP*TP*CP*AP*GP*TP*CP*TP*AP*GP*AP*CP*AP*TP*A)-3', 5'-D(P*GP*TP*AP*TP*GP*TP*CP*TP*AP*GP*AP*CP*TP*GP*A)-3', GLYCEROL, ... | | Authors: | Baburajendran, N, Palasingam, P, Narasimhan, K, Jauch, R, Kolatkar, P.R. | | Deposit date: | 2009-11-11 | | Release date: | 2010-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Smad1 MH1/DNA complex reveals distinctive rearrangements of BMP and TGF-beta effectors.
Nucleic Acids Res., 38, 2010
|
|
3KPY
 
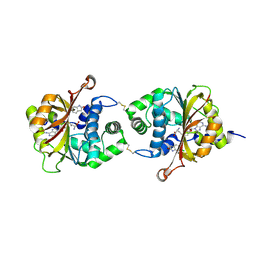 | | Crystal Structure of hPNMT in Complex AdoHcy and 6-Chlorooxindole | | Descriptor: | 6-chloro-1,3-dihydro-2H-indol-2-one, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
3KDI
 
 | | Structure of (+)-ABA bound PYL2 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Putative uncharacterized protein At2g26040 | | Authors: | Yin, P, Fan, H, Hao, Q, Yuan, X, Yan, N. | | Deposit date: | 2009-10-22 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.379 Å) | | Cite: | Structural insights into the mechanism of abscisic acid signaling by PYL proteins
Nat.Struct.Mol.Biol., 16, 2009
|
|
3KSC
 
 | | Crystal structure of pea prolegumin, an 11S seed globulin from Pisum sativum L. | | Descriptor: | GLYCEROL, LegA class, SULFATE ION | | Authors: | Tandang-Silvas, M.R.G, Fukuda, T, Fukuda, C, Prak, K, Cabanos, C, Kimura, A, Itoh, T, Mikami, B, Maruyama, N, Utsumi, S. | | Deposit date: | 2009-11-21 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Conservation and divergence on plant seed 11S globulins based on crystal structures.
Biochim.Biophys.Acta, 1804, 2010
|
|
3KJZ
 
 | | Crystal structure of native peptidyl-tRNA hydrolase from Mycobacterium smegmatis | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Kumar, A, Singh, N, Yadav, R, Prem Kumar, R, Sharma, S, Arora, A, Singh, T.P. | | Deposit date: | 2009-11-04 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of peptidyl-tRNA hydrolase from mycobacterium smegmatis reveals novel features related to enzyme dynamics.
Int J Biochem Mol Biol, 3, 2012
|
|
3KMS
 
 | | G62S mutant of foot-and-mouth disease virus RNA-polymerase in complex with a template- primer RNA trigonal structure | | Descriptor: | 3D polymerase, MAGNESIUM ION, RNA (5'-R(*AP*UP*GP*GP*GP*CP*C)-3'), ... | | Authors: | Ferrer-Orta, C, Verdaguer, N, Perez-Luque, R. | | Deposit date: | 2009-11-11 | | Release date: | 2010-07-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of foot-and-mouth disease virus mutant polymerases with reduced sensitivity to ribavirin
J.Virol., 84, 2010
|
|
3KQV
 
 | | Crystal Structure of hPNMT in Complex AdoHcy and Formanilide | | Descriptor: | FORMANILIDE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
5H3Z
 
 | | Crystal Structure of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Nakajima, M, Tanaka, N, Furukawa, N, Nihira, T, Kodutsumi, Y, Takahashi, Y, Sugimoto, N, Miyanaga, A, Fushinobu, S, Taguchi, H, Nakai, H. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic insight into the substrate specificity of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans
Sci Rep, 7, 2017
|
|
5H42
 
 | | Crystal Structure of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans in complex with alpha-d-glucose-1-phosphate | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, Uncharacterized protein, alpha-D-glucopyranose | | Authors: | Nakajima, M, Tanaka, N, Furukawa, N, Nihira, T, Kodutsumi, Y, Takahashi, Y, Sugimoto, N, Miyanaga, A, Fushinobu, S, Taguchi, H, Nakai, H. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanistic insight into the substrate specificity of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans
Sci Rep, 7, 2017
|
|
6S4A
 
 | | Structure of human MTHFD2 in complex with TH9028 | | Descriptor: | (2~{S})-2-[[5-[[2,4-bis(azanyl)-6-oxidanylidene-5~{H}-pyrimidin-5-yl]carbamoylamino]pyridin-2-yl]carbonylamino]-4-(1~{H}-1,2,3,4-tetrazol-5-yl)butanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, ... | | Authors: | Gustafsson, R, Scaletti, E.R, Bonagas, N, Gustafsson, N.M, Henriksson, M, Abdurakhmanov, E, Andersson, Y, Bengtsson, C, Borhade, S, Desroses, M, Farnegardh, K, Garg, N, Gokturk, C, Haraldsson, M, Iliev, P, Jarvius, M, Jemth, A.S, Kalderen, C, Karsten, S, Klingegard, F, Koolmeister, T, Martens, U, Llona-Minguez, S, Loseva, O, Marttila, P, Michel, M, Moulson, R, Nordstrom, H, Paulin, C, Pham, T, Pudelko, L, Rasti, A, Roos, A.K, Sarno, A, Sandberg, L, Scobie, M, Sjoberg, B, Svensson, R, Unterlass, J.E, Vallin, K, Vo, D, Wiita, E, Warpman-Berglund, U, Homan, E.J, Helleday, T, Stenmark, P. | | Deposit date: | 2019-06-27 | | Release date: | 2021-07-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pharmacological targeting of MTHFD2 suppresses acute myeloid leukemia by inducing thymidine depletion and replication stress.
Nat Cancer, 3, 2022
|
|
6S4F
 
 | | Structure of human MTHFD2 in complex with TH9619 | | Descriptor: | (E,4S)-4-[[5-[2-[2,6-bis(azanyl)-4-oxidanylidene-1H-pyrimidin-5-yl]ethanoylamino]-3-fluoranyl-pyridin-2-yl]carbonylamino]pent-2-enedioic acid, ADENOSINE-5'-DIPHOSPHATE, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, ... | | Authors: | Scaletti, E.R, Gustafsson, R, Bonagas, N, Gustafsson, N.M, Henriksson, M, Abdurakhmanov, E, Andersson, Y, Bengtsson, C, Borhade, S, Desroses, M, Farnegardh, K, Garg, N, Gokturk, C, Haraldsson, M, Iliev, P, Jarvius, M, Jemth, A.S, Kalderen, C, Karsten, S, Klingegard, F, Koolmeister, T, Martens, U, Llona-Minguez, S, Loseva, O, Marttila, P, Michel, M, Moulson, R, Nordstrom, H, Paulin, C, Pham, T, Pudelko, L, Rasti, A, Roos, A.K, Sarno, A, Sandberg, L, Scobie, M, Sjoberg, B, Svensson, R, Unterlass, J.E, Vallin, K, Vo, D, Wiita, E, Warpman-Berglund, U, Homan, E.J, Helleday, T, Stenmark, P. | | Deposit date: | 2019-06-27 | | Release date: | 2021-07-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Pharmacological targeting of MTHFD2 suppresses acute myeloid leukemia by inducing thymidine depletion and replication stress.
Nat Cancer, 3, 2022
|
|
6S4E
 
 | | Structure of human MTHFD2 in complex with TH7299 | | Descriptor: | (2S)-2-[[4-[[2,4-bis(azanyl)-6-oxidanylidene-1H-pyrimidin-5-yl]carbamoylamino]phenyl]carbonylamino]pentanedioic acid, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial, ... | | Authors: | Gustafsson, R, Scaletti, E.R, Bonagas, N, Gustafsson, N.M, Henriksson, M, Abdurakhmanov, E, Andersson, Y, Bengtsson, C, Borhade, S, Desroses, M, Farnegardh, K, Garg, N, Gokturk, C, Haraldsson, M, Iliev, P, Jarvius, M, Jemth, A.S, Kalderen, C, Karsten, S, Klingegard, F, Koolmeister, T, Martens, U, Llona-Minguez, S, Loseva, O, Marttila, P, Michel, M, Moulson, R, Nordstrom, H, Paulin, C, Pham, T, Pudelko, L, Rasti, A, Roos, A.K, Sarno, A, Sandberg, L, Scobie, M, Sjoberg, B, Svensson, R, Unterlass, J.E, Vallin, K, Vo, D, Wiita, E, Warpman-Berglund, U, Homan, E.J, Helleday, T, Stenmark, P. | | Deposit date: | 2019-06-27 | | Release date: | 2021-07-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pharmacological targeting of MTHFD2 suppresses acute myeloid leukemia by inducing thymidine depletion and replication stress.
Nat Cancer, 3, 2022
|
|
2RUH
 
 | | Chemical Shift Assignments for MIP and MDM2 in bound state | | Descriptor: | E3 ubiquitin-protein ligase Mdm2 | | Authors: | Nagata, T, Shirakawa, K, Kobayashi, N, Shiheido, H, Horisawa, K, Katahira, M, Doi, N, Yanagawa, H. | | Deposit date: | 2014-06-03 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Inhibition of the MDM2:p53 Interaction by an Optimized MDM2-Binding Peptide Selected with mRNA Display
Plos One, 9, 2014
|
|
8F0O
 
 | |
2RUQ
 
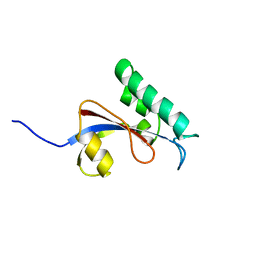 | | solution structure of human Pin1 PPIase mutant C113A | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Jing, W, Tochio, N, Tate, S. | | Deposit date: | 2015-01-20 | | Release date: | 2016-01-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Allosteric Breakage of the Hydrogen Bond within the Dual-Histidine Motif in the Active Site of Human Pin1 PPIase
Biochemistry, 54, 2015
|
|
2RRF
 
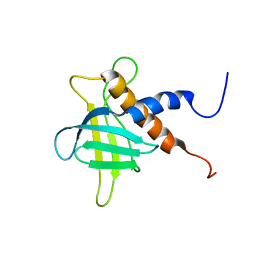 | | The solution structure of the C-terminal region of Zinc finger FYVE domain-containing protein 21 | | Descriptor: | Zinc finger FYVE domain-containing protein 21 | | Authors: | Koshiba, S, Tomizawa, T, Hayashi, F, Tochio, N, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S. | | Deposit date: | 2010-08-03 | | Release date: | 2011-08-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | ZF21 protein, a regulator of the disassembly of focal adhesions and cancer metastasis, contains a novel noncanonical pleckstrin homology domain
J.Biol.Chem., 286, 2011
|
|
2RUR
 
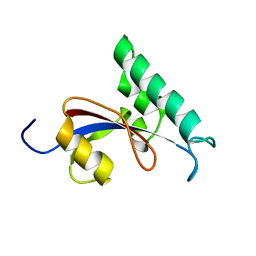 | | Solution structure of Human Pin1 PPIase C113S mutant | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Jing, W, Tochio, N, Tate, S. | | Deposit date: | 2015-01-20 | | Release date: | 2016-01-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Allosteric Breakage of the Hydrogen Bond within the Dual-Histidine Motif in the Active Site of Human Pin1 PPIase
Biochemistry, 54, 2015
|
|
8SLO
 
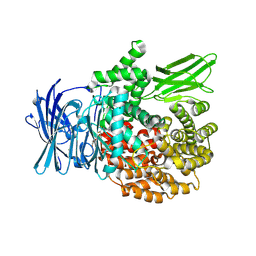 | | Plasmodium falciparum M1 aminopeptidase bound to selective inhibitor MIPS2673 | | Descriptor: | 2-hydroxy-N-[(1R)-2-(hydroxyamino)-2-oxo-1-(3',4',5'-trifluoro[1,1'-biphenyl]-4-yl)ethyl]-2-methylpropanamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | McGowan, S.M, Drinkwater, N. | | Deposit date: | 2023-04-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Chemoproteomics validates selective targeting of Plasmodium M1 alanyl aminopeptidase as an antimalarial strategy.
Elife, 13, 2024
|
|
