8YWO
 
 | | Crystal structure of L-azetidine-2-carboxylate hydrolase soaked in (S)-azetidine-2-carboxylic acid | | Descriptor: | (2S)-azetidine-2-carboxylic acid, (S)-2-haloacid dehalogenase | | Authors: | Toyoda, M, Mizutani, K, Mikami, B, Wackett, L.P, Esaki, N, Kurihara, T. | | Deposit date: | 2024-03-31 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Research for the crystal structure of L-azetidine-2-carboxylate hydrolase
To Be Published
|
|
5LZP
 
 | | Binding of the C-terminal GQYL motif of the bacterial proteasome activator Bpa to the 20S proteasome | | Descriptor: | Bacterial proteasome activator, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Bolten, M, Delley, C.L, Leibundgut, M, Boehringer, D, Ban, N, Weber-Ban, E. | | Deposit date: | 2016-09-30 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Analysis of the Bacterial Proteasome Activator Bpa in Complex with the 20S Proteasome.
Structure, 24, 2016
|
|
8YVW
 
 | | Crystal structure of D12N mutant of L-azetidine-2-carboxylate hydrolase | | Descriptor: | (S)-2-haloacid dehalogenase, FORMIC ACID, IMIDAZOLE, ... | | Authors: | Toyoda, M, Mizutani, K, Mikami, B, Wackett, L.P, Esaki, N, Kurihara, T. | | Deposit date: | 2024-03-29 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Research for the crystal structure of L-azetidine-2-carboxylate hydrolase
To Be Published
|
|
8YLB
 
 | | Cocrystal structures of agonists compound 1 with HsClpP | | Descriptor: | 5-[(2-methylphenyl)methyl]-11-(phenylmethyl)-2,5,7,11-tetrazatricyclo[7.4.0.0^{2,6}]trideca-1(9),6-dien-8-one, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | Authors: | Zhao, N, Zhu, Y, Bao, R. | | Deposit date: | 2024-03-06 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Rational Design of a Novel Class of Human ClpP Agonists through a Ring-Opening Strategy with Enhanced Antileukemia Activity.
J.Med.Chem., 67, 2024
|
|
5M06
 
 | | Crystal structure of Mycobacterium tuberculosis PknI kinase domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Serine/threonine-protein kinase PknI | | Authors: | Wagner, T, Lisa, M.N, Alexandre, M, Barilone, N, Raynal, B, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2016-10-03 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of PknI from Mycobacterium tuberculosis shows an inactive, pseudokinase-like conformation.
FEBS J., 284, 2017
|
|
5M07
 
 | | Crystal structure of Mycobacterium tuberculosis PknI kinase domain, C20A mutant | | Descriptor: | SODIUM ION, Serine/threonine-protein kinase PknI | | Authors: | Lisa, M.N, Wagner, T, Alexandre, M, Barilone, N, Raynal, B, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2016-10-03 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of PknI from Mycobacterium tuberculosis shows an inactive, pseudokinase-like conformation.
FEBS J., 284, 2017
|
|
5M08
 
 | | Crystal structure of Mycobacterium tuberculosis PknI kinase domain, C20A_R136A double mutant | | Descriptor: | Serine/threonine-protein kinase PknI | | Authors: | Lisa, M.N, Wagner, T, Alexandre, M, Barilone, N, Raynal, B, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2016-10-03 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | The crystal structure of PknI from Mycobacterium tuberculosis shows an inactive, pseudokinase-like conformation.
FEBS J., 284, 2017
|
|
5M09
 
 | | Crystal structure of Mycobacterium tuberculosis PknI kinase domain, C20A_R136N double mutant | | Descriptor: | SODIUM ION, Serine/threonine-protein kinase PknI | | Authors: | Lisa, M.N, Wagner, T, Alexandre, M, Barilone, N, Raynal, B, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2016-10-03 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | The crystal structure of PknI from Mycobacterium tuberculosis shows an inactive, pseudokinase-like conformation.
FEBS J., 284, 2017
|
|
5LUR
 
 | | An avidin mutant | | Descriptor: | Avidin, CHLORIDE ION, PROGESTERONE | | Authors: | Agrawal, N, Lehtonen, S.I, Riihimaki, T.A, Kulomaa, M.S, Hytonen, V.P, Johnson, M.S, Airenne, T.T. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An avidin mutant
TO BE PUBLISHED
|
|
5LOL
 
 | | Glutathione-bound Dehydroascorbate Reductase 2 of Arabidopsis thaliana | | Descriptor: | GLUTATHIONE, GLYCEROL, Glutathione S-transferase DHAR2, ... | | Authors: | Young, D.R, Pallo, A, Bodra, N, Messens, J. | | Deposit date: | 2016-08-09 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Arabidopsis thaliana dehydroascorbate reductase 2: Conformational flexibility during catalysis.
Sci Rep, 7, 2017
|
|
8ZN2
 
 | | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.65 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHONOACETIC ACID, ... | | Authors: | Ahmad, N, Sharma, P, Bhushan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.65 A resolution.
To Be Published
|
|
5LLM
 
 | | Structure of the thermostabilized EAAT1 cryst mutant in complex with L-ASP and the allosteric inhibitor UCPH101 | | Descriptor: | 2-Amino-5,6,7,8-tetrahydro-4-(4-methoxyphenyl)-7-(naphthalen-1-yl)-5-oxo-4H-chromene-3-carbonitrile, ASPARTIC ACID, Excitatory amino acid transporter 1,Neutral amino acid transporter B(0),Excitatory amino acid transporter 1, ... | | Authors: | Canul-Tec, J, Assal, R, Legrand, P, Reyes, N. | | Deposit date: | 2016-07-27 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure and allosteric inhibition of excitatory amino acid transporter 1.
Nature, 544, 2017
|
|
8ZN3
 
 | | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.41 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHONOACETIC ACID, ... | | Authors: | Ahmad, N, Sharma, P, Bhushan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.41 A resolution.
To Be Published
|
|
5LZD
 
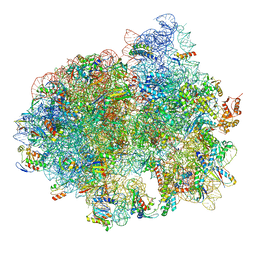 | | Structure of SelB-Sec-tRNASec bound to the 70S ribosome in the GTPase activated state (GA) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
5M1X
 
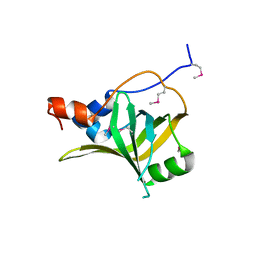 | | Crystal structure of S. cerevisiae Rfa1 N-OB domain mutant (K45E) | | Descriptor: | Replication factor A protein 1 | | Authors: | Seeber, A, Hegnauer, A.M, Hustedt, N, Deshpande, I, Poli, J, Eglinger, J, Pasero, P, Gut, H, Shinohara, M, Hopfner, K.P, Shimada, K, Gasser, S.M. | | Deposit date: | 2016-10-11 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | RPA Mediates Recruitment of MRX to Forks and Double-Strand Breaks to Hold Sister Chromatids Together.
Mol. Cell, 64, 2016
|
|
5LWP
 
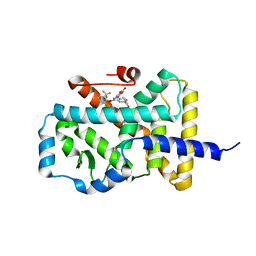 | | Discovery of phenoxyindazoles and phenylthioindazoles as RORg inverse agonists | | Descriptor: | 4-[3-[2-chloranyl-6-(trifluoromethyl)phenoxy]-5-(dimethylcarbamoyl)indazol-1-yl]benzoic acid, Nuclear receptor ROR-gamma | | Authors: | Ouvry, G, Bouix-Peter, C, Ciesielski, F, Chantalat, L, Christin, O, Comino, C, Duvert, D, Feret, C, Harris, C.S, Luzy, A.-P, Musicki, B, Orfila, D, Pascau, J, Parnet, V, Perrin, A, Pierre, R, Raffin, C, Rival, Y, Taquet, N, Thoreau, E, Hennequin, L.F. | | Deposit date: | 2016-09-19 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of phenoxyindazoles and phenylthioindazoles as ROR gamma inverse agonists.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5LXN
 
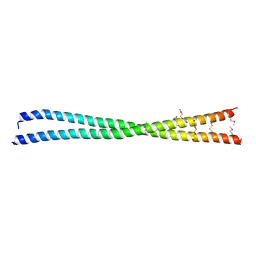 | | Coiled-coil protein | | Descriptor: | SULFATE ION, TRIETHYLENE GLYCOL, Transforming acidic coiled-coil-containing protein 3 | | Authors: | Thiyagarajan, N, Bunney, T.D, Katan, M. | | Deposit date: | 2016-09-22 | | Release date: | 2017-10-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Coiled-coil protein
To Be Published
|
|
5Z5F
 
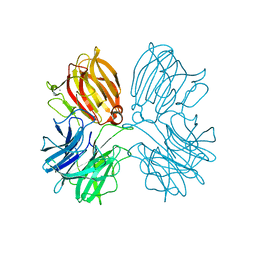 | | Crystal structure of a thermostable glycoside hydrolase family 43 {beta}-1,4-xylosidase from Geobacillus thermoleovorans IT-08 in complex with L-arabinose | | Descriptor: | Beta-xylosidase, CALCIUM ION, beta-L-arabinofuranose | | Authors: | Rohman, A, van Oosterwijk, N, Puspaningsih, N.N.T, Dijkstra, B.W. | | Deposit date: | 2018-01-18 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of product inhibition by arabinose and xylose of the thermostable GH43 beta-1,4-xylosidase from Geobacillus thermoleovorans IT-08.
PLoS ONE, 13, 2018
|
|
5LZE
 
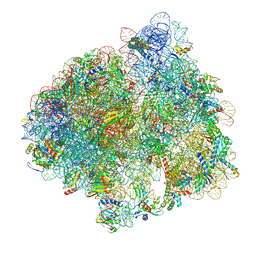 | | Structure of the 70S ribosome with Sec-tRNASec in the classical pre-translocation state (C) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
5LZC
 
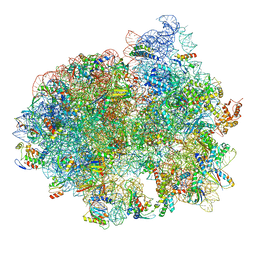 | | Structure of SelB-Sec-tRNASec bound to the 70S ribosome in the codon reading state (CR) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
5LRT
 
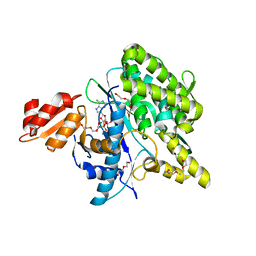 | | Structure of the Deamidase-Depupylase Dop of the Prokaryotic Ubiquitin-like Modification Pathway in Complex with ADP and Phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, Depupylase, ... | | Authors: | Bolten, M, Vahlensieck, C, Lipp, C, Leibundgut, M, Ban, N, Weber-Ban, E. | | Deposit date: | 2016-08-19 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Depupylase Dop Requires Inorganic Phosphate in the Active Site for Catalysis.
J. Biol. Chem., 292, 2017
|
|
5LWF
 
 | | Structure of a single domain camelid antibody fragment cAb-G10S in complex with the BlaP beta-lactamase from Bacillus licheniformis | | Descriptor: | ACETATE ION, Beta-lactamase, Camelid heavy-chain antibody variable fragment cAb-G10S | | Authors: | Vettore, N, Kerff, F, Pain, C, Herman, R, Sauvage, E, Preumont, S, Charlier, P, Dumoulin, M. | | Deposit date: | 2016-09-16 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Probing the mechanism of aggregation of polyQ model proteins with camelid heavy-chain antibody fragments
To Be Published
|
|
8Z3S
 
 | | Activation mechanism and novel binding site of the BKCa channel activator CTIBD | | Descriptor: | 4-[4-(4-chlorophenyl)-3-(trifluoromethyl)-1,2-oxazol-5-yl]benzene-1,3-diol, CALCIUM ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Lee, N, Kim, S, Jo, H, Lee, N.Y, Jin, M.S, Park, C.S. | | Deposit date: | 2024-04-16 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Activation mechanism and novel binding sites of the BK Ca channel activator CTIBD.
Life Sci Alliance, 7, 2024
|
|
4E81
 
 | |
5YWE
 
 | | Crystal structure of hematopoietic prostaglandin D synthase apo form | | Descriptor: | GLUTATHIONE, GLYCEROL, Hematopoietic prostaglandin D synthase, ... | | Authors: | Kamo, M, Furubayashi, N, Inaka, K, Aritake, K, Urade, Y. | | Deposit date: | 2017-11-29 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of hematopoietic prostaglandin D synthase apo form
To Be Published
|
|
