7NG4
 
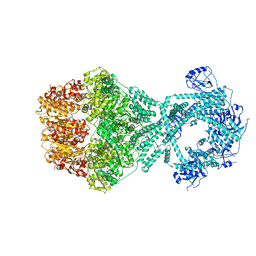 | | P1b-state of wild type human mitochondrial LONP1 protease with bound endogenous substrate protein and in presence of ATP/ADP mix | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Lon protease homolog, ... | | Authors: | Mohammed, I, Schmitz, K.A, Schenck, N, Maier, T, Abrahams, J.P. | | Deposit date: | 2021-02-08 | | Release date: | 2021-02-24 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Catalytic cycling of human mitochondrial Lon protease.
Structure, 30, 2022
|
|
5MAQ
 
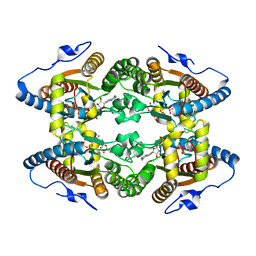 | | Crystal Structure of Polyphosphate Kinase from Meiothermus ruber bound to ADP and PPi | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PYROPHOSPHATE, ... | | Authors: | Gerhardt, S, Einsle, O, Kemper, F, Schwarzer, N. | | Deposit date: | 2016-11-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Substrate recognition and mechanism revealed by ligand-bound polyphosphate kinase 2 structures.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7NBY
 
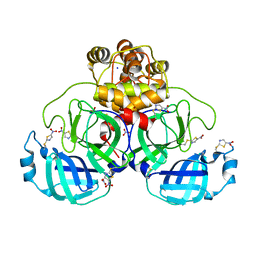 | | Crystal structure of SU3327 (halicin) covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | Descriptor: | 5-nitro-1,3-thiazole, CHLORIDE ION, Main Protease, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2021-01-28 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of SU3327 (halicin) covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2.
To Be Published
|
|
4ADF
 
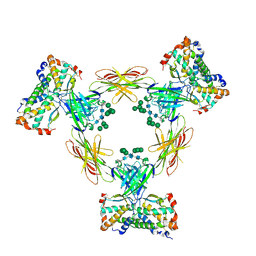 | | CRYSTAL STRUCTURE OF THE HUMAN COLONY-STIMULATING FACTOR 1 (hCSF-1) CYTOKINE IN COMPLEX WITH THE VIRAL RECEPTOR BARF1 | | Descriptor: | MACROPHAGE COLONY-STIMULATING FACTOR 1, SECRETED PROTEIN BARF1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Elegheert, J, Bracke, N, Savvides, S.N. | | Deposit date: | 2011-12-23 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Allosteric Competitive Inactivation of Hematopoietic Csf-1 Signaling by the Viral Decoy Receptor Barf1.
Nat.Struct.Mol.Biol., 19, 2012
|
|
7NEJ
 
 | | Solution structure of DNA:RNA hybrid duplex | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*AP*CP*GP*G)-3'), RNA (5'-R(*CP*CP*GP*UP*GP*UP*AP*G)-3') | | Authors: | Cabrero, C, Martin-Pintado, N, Mazzini, S, Gargallo, R, Eritja, R, Avino, A, Gonzalez, C. | | Deposit date: | 2021-02-04 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Effects of Incorporation of 2 -Deoxy-2 2 -difluorodeoxycytidine (Gemcitabine) in A- and B-form Duplexes.
Chemistry, 2021
|
|
7NG6
 
 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in spacegroup P1 in absence of DTT. | | Descriptor: | 3C-like proteinase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2021-02-08 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7NBN
 
 | | Allostery through DNA drives phenotype switching | | Descriptor: | AddAB promoter | | Authors: | Rosenblum, G, Elad, N, Rozenberg, H, Wiggers, F, Jungwirth, J, Hofmann, H. | | Deposit date: | 2021-01-27 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Allostery through DNA drives phenotype switching.
Nat Commun, 12, 2021
|
|
7NG3
 
 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in spacegroup P1. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2021-02-08 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7NGC
 
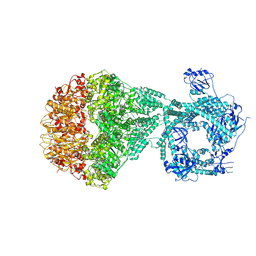 | | P2a-state of wild type human mitochondrial LONP1 protease with bound substrate protein and in presence of ATPgS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial, ... | | Authors: | Mohammed, I, Schmitz, K.A, Schenck, N, Maier, T, Abrahams, J.P. | | Deposit date: | 2021-02-09 | | Release date: | 2021-04-07 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Catalytic cycling of human mitochondrial Lon protease.
Structure, 30, 2022
|
|
7NBK
 
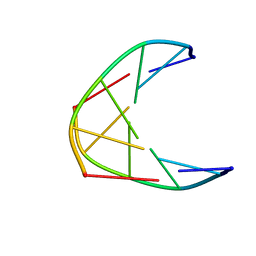 | | Solution structure of DNA duplex containing a 2'-deoxy-2'2'-difluorodeoxycytidine (gemcitabine) modification | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*(FFC)P*G)-3') | | Authors: | Cabrero, C, Martin-Pintado, N, Mazzini, S, Gargallo, R, Eritja, R, Avino, A, Gonzalez, C. | | Deposit date: | 2021-01-27 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Effects of Incorporation of 2 -Deoxy-2 2 -difluorodeoxycytidine (Gemcitabine) in A- and B-form Duplexes.
Chemistry, 2021
|
|
7NNS
 
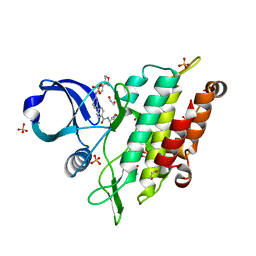 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound Momelotinib | | Descriptor: | 1,2-ETHANEDIOL, Activin receptor type I, Momelotinib, ... | | Authors: | Williams, E, Chen, Z, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound Momelotinib
To Be Published
|
|
7NF5
 
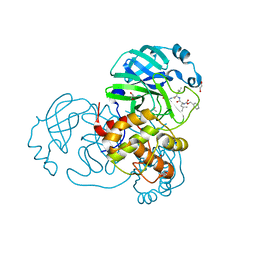 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in spacegroup C2. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2021-02-05 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7NBL
 
 | | Solution structure of DNA:RNA hybrid containing a 2'-deoxy-2'2'-difluorodeoxycytidine (gemcitabine) modification | | Descriptor: | DNA (5'-D(*CP*TP*AP*(FFC)P*AP*CP*GP*G)-3'), RNA (5'-R(*CP*CP*GP*UP*GP*UP*AP*G)-3') | | Authors: | Cabrero, C, Martin-Pintado, N, Mazzini, S, Gargallo, R, Eritja, R, Avino, A, Gonzalez, C. | | Deposit date: | 2021-01-27 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Effects of Incorporation of 2 -Deoxy-2 2 -difluorodeoxycytidine (Gemcitabine) in A- and B-form Duplexes.
Chemistry, 2021
|
|
7NO8
 
 | | Structure of the mature RSV CA lattice: Group I, pentamer-hexamer interface, class 1"6 | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOM
 
 | | Structure of the mature RSV CA lattice: Group IV, hexamer-hexamer interface, class 4'Beta | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NO1
 
 | | Structure of the mature RSV CA lattice: T=3 CA icosahedron | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOE
 
 | | Structure of the mature RSV CA lattice: Group III, hexamer-hexamer interface, class 3'5 | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NO2
 
 | | Structure of the mature RSV CA lattice: hexamer derived from tubes (C2-symmetric) | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOD
 
 | | Structure of the mature RSV CA lattice: Group III, hexamer-hexamer interface, class 3'4 | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOG
 
 | | Structure of the mature RSV CA lattice: Group III, hexamer-hexamer interface, class 4'5 | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NO9
 
 | | Structure of the mature RSV CA lattice: Group I, pentamer-pentamer interface, class 1'1 | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NO0
 
 | | Structure of the mature RSV CA lattice: T=1 CA icosahedron | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOO
 
 | | Structure of the mature RSV CA lattice: Group IV, hexamer-hexamer interface, class 5'Alpha | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NO5
 
 | | Structure of the mature RSV CA lattice: hexamer with 2 adjacent pentamers (C2 symmetric) | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOB
 
 | | Structure of the mature RSV CA lattice: Group II, hexamer-hexamer interface, class 2'6 | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
