2GVF
 
 | | HCV NS3-4A protease domain complexed with a macrocyclic ketoamide inhibitor, SCH419021 | | Descriptor: | (6R,8S,11S)-11-CYCLOHEXYL-N-(1-{[(2-{[(1S)-2-(DIMETHYLAMINO)-2-OXO-1-PHENYLETHYL]AMINO}-2-OXOETHYL)AMINO](OXO)ACETYL}BUTYL)-10,13-DIOXO-2,5-DIOXA-9,12-DIAZATRICYCLO[14.3.1.1~6,9~]HENICOSA-1(20),16,18-TRIENE-8-CARBOXAMIDE, Polyprotein, ZINC ION, ... | | Authors: | Arasappan, A, Njoroge, F.G, Chen, K.X, Venkatraman, S, Parekh, T.N, Gu, H, Pichardo, J, Butkiewicz, N, Prongay, A, Madison, V, Girijavallabhan, V. | | Deposit date: | 2006-05-02 | | Release date: | 2007-01-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | P2-P4 macrocyclic inhibitors of hepatitis C virus NS3-4A serine protease.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1DIO
 
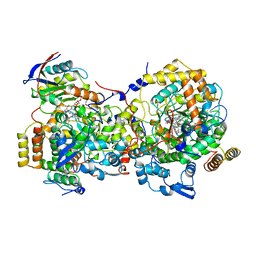 | | DIOL DEHYDRATASE-CYANOCOBALAMIN COMPLEX FROM KLEBSIELLA OXYTOCA | | Descriptor: | COBALAMIN, POTASSIUM ION, PROTEIN (DIOL DEHYDRATASE), ... | | Authors: | Shibata, N, Masuda, J, Tobimatsu, T, Toraya, T, Suto, K, Morimoto, Y, Yasuoka, N. | | Deposit date: | 1999-01-27 | | Release date: | 2000-01-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A new mode of B12 binding and the direct participation of a potassium ion in enzyme catalysis: X-ray structure of diol dehydratase.
Structure Fold.Des., 7, 1999
|
|
5X2Q
 
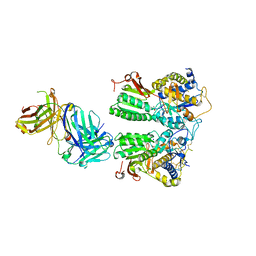 | | Crystal structure of the medaka fish taste receptor T1r2a-T1r3 ligand binding domains in complex with glycine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Nuemket, N, Yasui, N, Atsumi, N, Yamashita, A. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for perception of diverse chemical substances by T1r taste receptors
Nat Commun, 8, 2017
|
|
6ZOP
 
 | | Structure of the cysteine-rich domain of PiggyMac, a domesticated PiggyBac transposase involved in programmed genome rearrangements | | Descriptor: | DDE_Tnp_1_7 domain-containing protein, ZINC ION | | Authors: | Bessa, L, Guerineau, M, Moriau, S, Lescop, E, Bontems, F, Mathy, N, Guittet, E, Bischerour, J, Betermier, M, Morellet, N. | | Deposit date: | 2020-07-07 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | SOLUTION NMR | | Cite: | The unusual structure of the PiggyMac cysteine-rich domain reveals zinc finger diversity in PiggyBac-related transposases.
Mob DNA, 12, 2021
|
|
6MHE
 
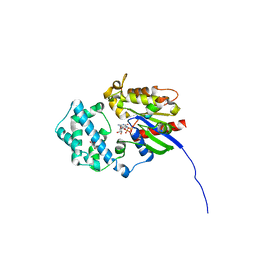 | | Galphai3 co-crystallized with KB752 | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(k) subunit alpha, ... | | Authors: | Rees, S.D, Kalogriopoulos, N.A, Ngo, T, Kopcho, N, Ilatovskiy, A, Sun, N, Komives, E, Chang, G, Ghosh, P, Kufareva, I. | | Deposit date: | 2018-09-17 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for GPCR-independent activation of heterotrimeric Gi proteins.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6MHF
 
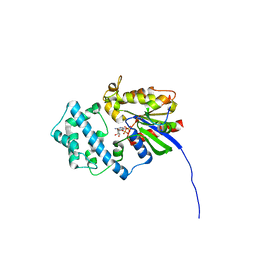 | | Galphai3 co-crystallized with GIV/Girdin | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, Girdin, ... | | Authors: | Rees, S.D, Kalogriopoulos, N.A, Ngo, T, Kopcho, N, Ilatovskiy, A, Sun, N, Komives, E, Chang, G, Ghosh, P, Kufareva, I. | | Deposit date: | 2018-09-17 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for GPCR-independent activation of heterotrimeric Gi proteins.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8UC1
 
 | |
7AMD
 
 | | In situ assembly of choline acetyltransferase ligands by a hydrothiolation reaction reveals key determinants for inhibitor design | | Descriptor: | Choline O-acetyltransferase, SODIUM ION, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3~{R})-2,2-dimethyl-4-[[3-[2-[(1~{R})-2-(1-methylpyridin-4-yl)-1-naphthalen-1-yl-ethyl]sulfanylethylamino]-3-oxidanylidene-propyl]amino]-3-oxidanyl-4-oxidanylidene-butyl] hydrogen phosphate | | Authors: | Allgardsson, A, Ekstrom, F.J, Wiktelius, D, Bergstrom, T, Hoster, N, Akfur, C, Forsgren, N, Lejon, C, Hedenstrom, M, Linusson, A. | | Deposit date: | 2020-10-08 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | In Situ Assembly of Choline Acetyltransferase Ligands by a Hydrothiolation Reaction Reveals Key Determinants for Inhibitor Design.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
8H1O
 
 | | Cryo-EM structure of KpFtsZ-monobody double helical tube | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Mb(Ec/KpFtsZ_S1) | | Authors: | Fujita, J, Amesaka, H, Yoshizawa, T, Kuroda, N, Kamimura, N, Hara, M, Inoue, T, Namba, K, Tanaka, S, Matsumura, H. | | Deposit date: | 2022-10-03 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
6JT3
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(4R,5R,6R)-2-amino-5-fluoro-4,6-dimethyl-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tadano, G, Komano, K, Yoshida, S, Suzuki, S, Nakahara, K, Fuchino, K, Fujimoto, K, Matsuoka, E, Yamamoto, T, Asada, N, Ito, H, Sakaguchi, G, Kanegawa, N, Kido, Y, Ando, S, Fukushima, T, Teisman, A, Urmaliya, V, Dhuyvetter, D, Borghys, H, Bergh, A.V.D, Austin, N, Gijsen, H.J.M, Yamano, Y, Iso, Y, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of an Extremely Potent Thiazine-Based beta-Secretase Inhibitor with Reduced Cardiovascular and Liver Toxicity at a Low Projected Human Dose.
J.Med.Chem., 62, 2019
|
|
5HO3
 
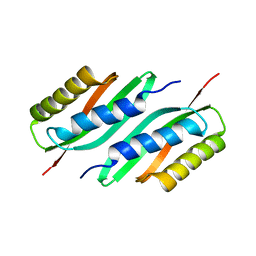 | | MamB | | Descriptor: | Magnetosome protein MamB | | Authors: | Zeytuni, N, Keren, N, Davidov, G, Zarivach, R. | | Deposit date: | 2016-01-19 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The dual role of MamB in magnetosome membrane assembly and magnetite biomineralization.
Mol. Microbiol., 107, 2018
|
|
8W86
 
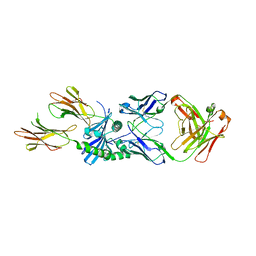 | | HLA-DQ2.5-B/C hordein peptide in complex with DQN0385AE02 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DQN0385AE02 Fab heavy chain, DQN0385AE02 Fab light chain, ... | | Authors: | Irie, M, Tsushima, T, Teranishi-Ikawa, Y, Takahashi, N, Ishii, S, Okura, Y, Fukami, T.A, Torizawa, T. | | Deposit date: | 2023-08-31 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.236 Å) | | Cite: | Characterizations of a neutralizing antibody broadly reactive to multiple gluten peptide:HLA-DQ2.5 complexes in the context of celiac disease.
Nat Commun, 14, 2023
|
|
8W85
 
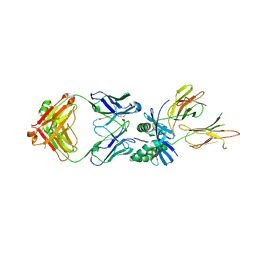 | | HLA-DQ2.5-gamma2 gliadin peptide in complex with DQN0385AE01 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DQN0385AE01 Fab heavy chain, DQN0385AE01 Fab light chain, ... | | Authors: | Irie, M, Tsushima, T, Teranishi-Ikawa, Y, Takahashi, N, Ishii, S, Okura, Y, Fukami, T.A, Torizawa, T. | | Deposit date: | 2023-08-31 | | Release date: | 2023-11-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.769 Å) | | Cite: | Characterizations of a neutralizing antibody broadly reactive to multiple gluten peptide:HLA-DQ2.5 complexes in the context of celiac disease.
Nat Commun, 14, 2023
|
|
8W84
 
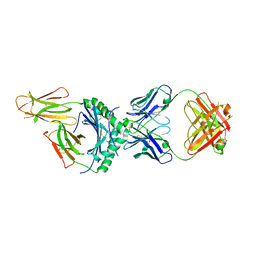 | | HLA-DQ2.5-alpha2 gliadin peptide in complex with DQN0344AE02 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DQN0344AE02 Fab heavy chain, DQN0344AE02 Fab light chain, ... | | Authors: | Irie, M, Tsushima, T, Teranishi-Ikawa, Y, Takahashi, N, Ishii, S, Okura, Y, Fukami, T.A, Torizawa, T. | | Deposit date: | 2023-08-31 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Characterizations of a neutralizing antibody broadly reactive to multiple gluten peptide:HLA-DQ2.5 complexes in the context of celiac disease.
Nat Commun, 14, 2023
|
|
8W83
 
 | | HLA-DQ2.5-alpha1 gliadin peptide in complex with DQN0344AE02 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DQN0344AE02 Fab heavy chain, DQN0344AE02 Fab light chain, ... | | Authors: | Irie, M, Tsushima, T, Teranishi-Ikawa, Y, Takahashi, N, Ishii, S, Okura, Y, Fukami, T.A, Torizawa, T. | | Deposit date: | 2023-08-31 | | Release date: | 2023-11-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.818 Å) | | Cite: | Characterizations of a neutralizing antibody broadly reactive to multiple gluten peptide:HLA-DQ2.5 complexes in the context of celiac disease.
Nat Commun, 14, 2023
|
|
8VQ1
 
 | | Pseudomonas fluorescens G150T isocyanide hydratase at 298 K XFEL data, thioimidate intermediate | | Descriptor: | CHLORIDE ION, Isonitrile hydratase InhA, N-(4-nitrophenyl)methanimine | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2024-01-17 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Changes in an enzyme ensemble during catalysis observed by high-resolution XFEL crystallography.
Sci Adv, 10, 2024
|
|
8VPW
 
 | | Pseudomonas fluorescens G150T isocyanide hydratase at 298 K XFEL data, free enzyme | | Descriptor: | CHLORIDE ION, Isonitrile hydratase InhA | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2024-01-17 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Changes in an enzyme ensemble during catalysis observed by high-resolution XFEL crystallography.
Sci Adv, 10, 2024
|
|
8IBL
 
 | | MES bound form of PET-degrading cutinase Cut190 with thermostability-improving mutations of S226P/R228S/Q138A/D250C-E296C/Q123H/N202H and S176A inactivation | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha/beta hydrolase family protein, CALCIUM ION, ... | | Authors: | Emori, M, Numoto, N, Kamiya, N, Oda, M. | | Deposit date: | 2023-02-10 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Improvement of thermostability and activity of PET-degrading enzyme Cut190 towards a detailed understanding and application of the enzymatic reaction mechanism.
Biorxiv, 2023
|
|
8IBM
 
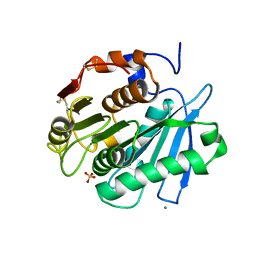 | | Sulfate bound form of PET-degrading cutinase Cut190 with thermostability-improving mutations of S226P/R228S/Q138A/D250C-E296C/Q123H/N202H and S176A inactivation | | Descriptor: | Alpha/beta hydrolase family protein, CALCIUM ION, SULFATE ION | | Authors: | Emori, M, Numoto, N, Kamiya, N, Oda, M. | | Deposit date: | 2023-02-10 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Improvement of thermostability and activity of PET-degrading enzyme Cut190 towards a detailed understanding and application of the enzymatic reaction mechanism.
Biorxiv, 2023
|
|
5HO5
 
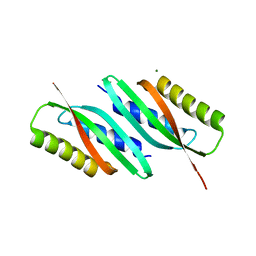 | | MamB | | Descriptor: | MAGNESIUM ION, Magnetosome protein MamB | | Authors: | Zeytuni, N, Keren, N, Davidov, G, Zarivach, R. | | Deposit date: | 2016-01-19 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The dual role of MamB in magnetosome membrane assembly and magnetite biomineralization.
Mol. Microbiol., 107, 2018
|
|
6SK7
 
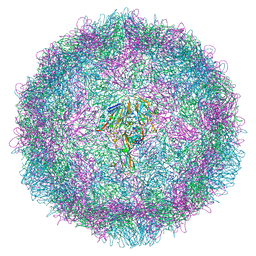 | | Cryo-EM structure of rhinovirus-A89 | | Descriptor: | VP1 capsid protein, VP2 capsid protein, VP3 capsid protein, ... | | Authors: | Wald, J, Goessweiner-Mohr, N, Blaas, D, Pasin, M. | | Deposit date: | 2019-08-14 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of pleconaril-resistant rhinovirus-B5 complexed to the antiviral OBR-5-340 reveals unexpected binding site.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8IB2
 
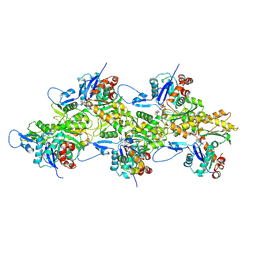 | | Structure of mammalian spectrin-actin junctional complex of membrane skeleton, Pointed-end segment, headpiece domain of dematin optimized | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Li, N, Chen, S, Gao, N. | | Deposit date: | 2023-02-09 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of membrane skeleton organization in red blood cells.
Cell, 186, 2023
|
|
8IC9
 
 | | Lys48-linked K48C-diubiquitin | | Descriptor: | Polyubiquitin-B, Ubiquitin | | Authors: | Hiranyakorn, M, Yagi-Utsumi, M, Yanaka, S, Ohtsuka, N, Momiyama, N, Satoh, T, Kato, K. | | Deposit date: | 2023-02-11 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Mutational and Environmental Effects on the Dynamic Conformational Distributions of Lys48-Linked Ubiquitin Chains.
Int J Mol Sci, 24, 2023
|
|
5HOK
 
 | | MamB-CTD mutant - D247A | | Descriptor: | MAGNESIUM ION, Magnetosome protein MamB | | Authors: | Keren, N, Zeytuni, N, Zarivach, R. | | Deposit date: | 2016-01-19 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The dual role of MamB in magnetosome membrane assembly and magnetite biomineralization.
Mol. Microbiol., 107, 2018
|
|
8IBN
 
 | | Cryo-EM structure of KpFtsZ single filament | | Descriptor: | Cell division protein FtsZ, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, POTASSIUM ION | | Authors: | Fujita, J, Amesaka, H, Yoshizawa, T, Kuroda, N, Kamimura, N, Hibino, K, Konishi, T, Kato, Y, Hara, M, Inoue, T, Namba, K, Tanaka, S, Matsumura, H. | | Deposit date: | 2023-02-10 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
