8VCZ
 
 | |
4PIS
 
 | | Crystal structure of human adenovirus 8 protease in complex with a nitrile inhibitor | | Descriptor: | N~2~-[(2R)-2-(3,5-dichlorophenyl)-2-(dimethylamino)acetyl]-N-({2-[(Z)-iminomethyl]pyrimidin-4-yl}methyl)-L-isoleucinamide, PVI, Protease | | Authors: | Mac Sweeney, A, Grosche, P, Ellis, D, Combrink, K, Erbel, P, Hughes, N, Sirockin, F, Melkko, S, Bernardi, A, Ramage, P, Jarousse, N, Altmann, E. | | Deposit date: | 2014-05-09 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and structure-based optimization of adenain inhibitors.
Acs Med.Chem.Lett., 5, 2014
|
|
8VD3
 
 | |
8VMG
 
 | | Crystal structure of GSK-3 26-383 bound to Axin 383-435 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Enos, M.D, Gavagan, M, Jameson, N, Zalatan, J.G, Weis, W.I. | | Deposit date: | 2024-01-13 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural and functional effects of phosphopriming and scaffolding in the kinase GSK-3 beta.
Sci.Signal., 17, 2024
|
|
8VCD
 
 | |
6BKD
 
 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2 core from genotype 6a bound to broadly neutralizing antibody AR3D | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab AR3D heavy chain, Fab AR3D light chain, ... | | Authors: | Tzarum, N, Wilson, I.A, Law, M. | | Deposit date: | 2017-11-08 | | Release date: | 2018-12-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Genetic and structural insights into broad neutralization of hepatitis C virus by human VH1-69 antibodies.
Sci Adv, 5, 2019
|
|
8VME
 
 | | Crystal structure of the GSK-3/Axin complex bound to a phosphorylated beta-catenin T41A peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Axin-1, Catenin beta-1, ... | | Authors: | Enos, M.D, Gavagan, M, Jameson, N, Zalatan, J.G, Weis, W.I. | | Deposit date: | 2024-01-13 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional effects of phosphopriming and scaffolding in the kinase GSK-3 beta.
Sci.Signal., 17, 2024
|
|
8VMF
 
 | | Crystal structure of a transition-state mimic of the GSK-3/Axin complex bound to a beta-catenin S45D peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Axin-1, ... | | Authors: | Enos, M.D, Gavagan, M, Jameson, N, Zalatan, J.G, Weis, W.I. | | Deposit date: | 2024-01-13 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional effects of phosphopriming and scaffolding in the kinase GSK-3 beta.
Sci.Signal., 17, 2024
|
|
6CHR
 
 | | Crystal structure of a group II intron lariat with an intact 3' splice site (pre-2s state) | | Descriptor: | MAGNESIUM ION, RNA (5'-R(P*UP*GP*UP*UP*UP*AP*UP*UP*AP*AP*AP*AP*A)-3'), RNA (621-MER), ... | | Authors: | Chan, R.T, Peters, J.K, Robart, A.R, Wiryaman, T, Rajashankar, K.R, Toor, N. | | Deposit date: | 2018-02-22 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for the second step of group II intron splicing.
Nat Commun, 9, 2018
|
|
8VTU
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with macrolone MCX-66, mRNA, aminoacylated A-site Phe-tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.40A resolution | | Descriptor: | 1-cyclopropyl-7-[(4-{[3-({(3aR,4R,7R,8S,9S,10R,11R,13R,14E,15S,15aR)-10-{[(2S,3R,4S,6R)-4-(dimethylamino)-3-hydroxy-6-methyloxan-2-yl]oxy}-4-ethyl-11-methoxy-3a,7,9,11,13,15-hexamethyl-14-[({3-[5-(methylcarbamoyl)pyridin-3-yl]prop-2-yn-1-yl}oxy)imino]-2,6-dioxododecahydro-2H,4H-[1,3]dioxolo[4,5-c]oxacyclotetradecin-8-yl}oxy)-3-oxopropyl]amino}butyl)amino]-6-fluoro-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Aleksandrova, E.V, Ma, C.-X, Klepacki, D, Alizadeh, F, Vazquez-Laslop, N, Liang, J.-H, Polikanov, Y.S, Mankin, A.S. | | Deposit date: | 2024-01-27 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Macrolones target bacterial ribosomes and DNA gyrase and can evade resistance mechanisms.
Nat.Chem.Biol., 2024
|
|
1VIE
 
 | |
4PIE
 
 | | Crystal structure of human adenovirus 2 protease a substrate based nitrile inhibitor | | Descriptor: | ACETATE ION, N-{(2S)-2-(3-chlorophenyl)-2-[(methylsulfonyl)amino]acetyl}-L-phenylalanyl-N-[(2Z)-2-iminoethyl]glycinamide, Pre-protein VI, ... | | Authors: | Mac Sweeney, A, Grosche, P, Ellis, D, Combrink, C, Erbel, C, Hughes, N, Sirockin, F, Melkko, S, Bernardi, A, Ramage, P, Jarousse, N, Altmann, E. | | Deposit date: | 2014-05-08 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery and structure-based optimization of adenain inhibitors.
Acs Med.Chem.Lett., 5, 2014
|
|
6DFP
 
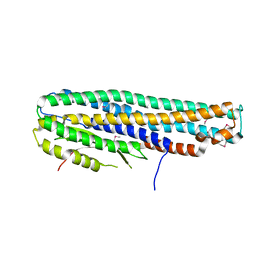 | | Crystal Structure of a Tripartite Toxin Component VCA0883 from Vibrio cholerae | | Descriptor: | VCA0883 | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-05-15 | | Release date: | 2018-05-23 | | Last modified: | 2022-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Genomic Island of Vibrio cholerae Encodes a Three-Component Cytotoxin with Monomer and Protomer Forms Structurally Similar to Alpha-Pore-Forming Toxins.
J.Bacteriol., 204, 2022
|
|
8WFM
 
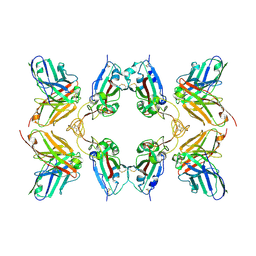 | |
8WG4
 
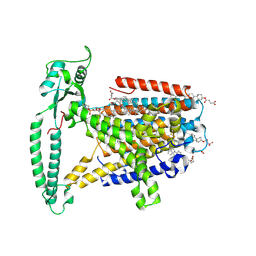 | | mouse TMEM63b in DDM-CHS micelle with YN9303-24 Fab | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHOLESTEROL HEMISUCCINATE, CSC1-like protein 2,Green fluorescent protein | | Authors: | Miyata, Y, Takahashi, K, Lee, Y, Sultan, C.S, Kuribayashi, R, Takahashi, M, Hata, K, Bamba, T, Izumi, Y, Liu, K, Uemura, T, Nomura, N, Iwata, S, Nagata, S, Nishizawa, T, Segawa, K. | | Deposit date: | 2023-09-20 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanosensitive channel TMEM63B functions as a plasma membrane lipid scramblase
To Be Published
|
|
6DBY
 
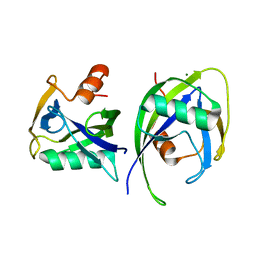 | | Crystal structure of Nudix 1 from Arabidopsis thaliana | | Descriptor: | MAGNESIUM ION, Nudix hydrolase 1 | | Authors: | Noel, J.P, Thomas, S.T, Dudareva, N, Henry, L.K. | | Deposit date: | 2018-05-03 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Contribution of isopentenyl phosphate to plant terpenoid metabolism.
Nat Plants, 4, 2018
|
|
1VIF
 
 | | STRUCTURE OF DIHYDROFOLATE REDUCTASE | | Descriptor: | DIHYDROFOLATE REDUCTASE, FOLIC ACID | | Authors: | Narayana, N, Matthews, D.A, Howell, E.E, Xuong, N.-H. | | Deposit date: | 1996-10-03 | | Release date: | 1997-10-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A plasmid-encoded dihydrofolate reductase from trimethoprim-resistant bacteria has a novel D2-symmetric active site.
Nat.Struct.Biol., 2, 1995
|
|
8WFH
 
 | |
6DCS
 
 | | Stage III sporulation protein AF (SpoIIIAF) | | Descriptor: | SULFATE ION, Stage III sporulation protein AF | | Authors: | Strynadka, N.C.J, Zeytuni, N, Camp, A.H, Flanagan, K.A. | | Deposit date: | 2018-05-08 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and biochemical characterization of SpoIIIAF, a component of a sporulation-essential channel in Bacillus subtilis.
J. Struct. Biol., 204, 2018
|
|
6DRA
 
 | | Low IP3 Ca2+ human type 3 1,4,5-inositol trisphosphate receptor | | Descriptor: | CALCIUM ION, Inositol 1,4,5-trisphosphate receptor type 3, ZINC ION | | Authors: | Hite, R.K, Paknejad, N. | | Deposit date: | 2018-06-11 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Structural basis for the regulation of inositol trisphosphate receptors by Ca2+and IP3.
Nat. Struct. Mol. Biol., 25, 2018
|
|
8WG3
 
 | | mouse TMEM63b in LMNG-CHS micelle | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHOLESTEROL HEMISUCCINATE, CSC1-like protein 2,Green fluorescent protein | | Authors: | Miyata, Y, Takahashi, K, Lee, Y, Sultan, C.S, Kuribayashi, R, Takahashi, M, Hata, K, Bamba, T, Izumi, Y, Liu, K, Uemura, T, Nomura, N, Iwata, S, Nagata, S, Nishizawa, T, Segawa, K. | | Deposit date: | 2023-09-20 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanosensitive channel TMEM63B functions as a plasma membrane lipid scramblase
To Be Published
|
|
6D58
 
 | | Crystal structure of a Fc fragment of Human IgG3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin heavy constant gamma 3 | | Authors: | Gohain, N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2018-04-19 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of a Fc fragment of Human IgG3.
To Be Published
|
|
6DMQ
 
 | |
6DO4
 
 | | KLHDC2 ubiquitin ligase in complex with SelS C-end degron | | Descriptor: | Kelch domain-containing protein 2, SELS C-END DEGRON | | Authors: | Rusnac, D.V, Lin, H.C, Yen, H.C.S, Zheng, N. | | Deposit date: | 2018-06-08 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Recognition of the Diglycine C-End Degron by CRL2KLHDC2Ubiquitin Ligase.
Mol. Cell, 72, 2018
|
|
6BV9
 
 | | Structure of proteinaceous RNase P 1 (PRORP1) from A. thaliana after overnight soak with juglone | | Descriptor: | 5-hydroxynaphthalene-1,4-dione, CHLORIDE ION, Proteinaceous RNase P 1, ... | | Authors: | Karasik, A, Wu, N, Fierke, C.A, Koutmos, M. | | Deposit date: | 2017-12-12 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of protein-only RNase P with Gambogic acid and Juglone
To Be Published
|
|
