6L5W
 
 | | Carbonmonoxy human hemoglobin C in the R quaternary structure at 140 K: Light (2 min) | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Shibayama, N, Park, S.Y, Ohki, M, Sato-Tomita, A. | | Deposit date: | 2019-10-24 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct observation of ligand migration within human hemoglobin at work.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1D79
 
 | | HIGH RESOLUTION REFINEMENT OF THE HEXAGONAL A-DNA OCTAMER D(GTGTACAC) AT 1.4 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*GP*TP*GP*TP*AP*CP*AP*C)-3') | | Authors: | Thota, N, Li, X.H, Bingman, C.A, Sundaralingam, M. | | Deposit date: | 1992-06-12 | | Release date: | 1993-04-15 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-resolution refinement of the hexagonal A-DNA octamer d(GTGTACAC) at 1.4 A.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
6L7V
 
 | |
1COA
 
 | | THE EFFECT OF CAVITY CREATING MUTATIONS IN THE HYDROPHOBIC CORE OF CHYMOTRYPSIN INHIBITOR 2 | | Descriptor: | CHYMOTRYPSIN INHIBITOR 2 | | Authors: | Jackson, S.E, Moracci, M, Elmasry, N, Johnson, C.M, Fersht, A.R. | | Deposit date: | 1993-05-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Effect of cavity-creating mutations in the hydrophobic core of chymotrypsin inhibitor 2.
Biochemistry, 32, 1993
|
|
6L5X
 
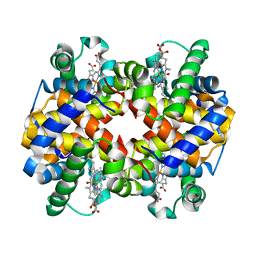 | | Carbonmonoxy human hemoglobin A in the R2 quaternary structure at 95 K: Light (2 min) | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Shibayama, N, Park, S.Y, Ohki, M, Sato-Tomita, A. | | Deposit date: | 2019-10-24 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Direct observation of ligand migration within human hemoglobin at work.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6XKU
 
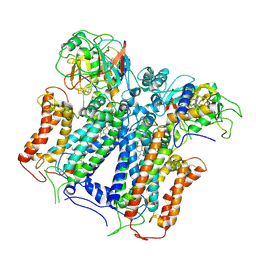 | | R. capsulatus cyt bc1 with one FeS protein in b position and one in c position (CIII2 b-c) | | Descriptor: | Cytochrome b, Cytochrome c1, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Steimle, S, Van Eeuwen, T, Ozturk, Y, Kim, H.J, Braitbard, M, Selamoglu, N, Garcia, B.A, Schneidman-Duhovny, D, Murakami, K, Daldal, F. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-30 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures of engineered active bc 1 -cbb 3 type CIII 2 CIV super-complexes and electronic communication between the complexes.
Nat Commun, 12, 2021
|
|
6XKZ
 
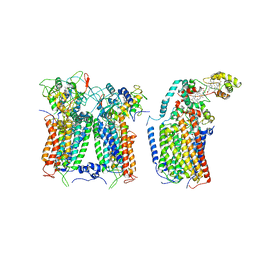 | | R. capsulatus CIII2CIV tripartite super-complex, conformation B (SC-1B) | | Descriptor: | COPPER (II) ION, Cbb3-type cytochrome c oxidase subunit CcoP,Cytochrome c-type cyt cy, Cytochrome b, ... | | Authors: | Steimle, S, Van Eeuwen, T, Ozturk, Y, Kim, H.J, Braitbard, M, Selamoglu, N, Garcia, B.A, Schneidman-Duhovny, D, Murakami, K, Daldal, F. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-30 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Cryo-EM structures of engineered active bc 1 -cbb 3 type CIII 2 CIV super-complexes and electronic communication between the complexes.
Nat Commun, 12, 2021
|
|
6X9G
 
 | |
6XL2
 
 | |
6L1V
 
 | | Domain-swapped Alcaligenes xylosoxidans azurin dimer | | Descriptor: | Azurin-1, COPPER (II) ION | | Authors: | Cahyono, R.N, Yamanaka, M, Nagao, S, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2019-09-30 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 3D domain swapping of azurin from Alcaligenes xylosoxidans.
Metallomics, 12, 2020
|
|
6KZO
 
 | | membrane protein | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yan, N. | | Deposit date: | 2019-09-25 | | Release date: | 2019-12-18 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of apo and antagonist-bound human Cav3.1.
Nature, 576, 2019
|
|
6XZH
 
 | | Structure of zVDR LBD-Calcitriol in complex with chimera 10 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, ARG-HIS-LYS-ILE-URL-URK-URL-LEU-GLN, Vitamin D3 receptor A | | Authors: | Buratto, J, Belorusova, A.Y, Rochel, N, Guichard, G. | | Deposit date: | 2020-02-04 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.372 Å) | | Cite: | Structural Basis for alpha-Helix Mimicry and Inhibition of Protein-Protein Interactions with Oligourea Foldamers.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6L2D
 
 | | Crystal structure of a cupin protein (tm1459) in copper (Cu) substituted form | | Descriptor: | COPPER (II) ION, Cupin_2 domain-containing protein | | Authors: | Fujieda, N, Ichihashi, H, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-10-03 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.198 Å) | | Cite: | Cupin Variants as a Macromolecular Ligand Library for Stereoselective Michael Addition of Nitroalkanes.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6X8W
 
 | |
6XZI
 
 | | Structure of zVDR LBD-calcitriol in complex with chimera 11 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, ACETATE ION, ARG-HIS-LYS-ILE-LEU-URK-UIL-URL, ... | | Authors: | Buratto, J, Belorusova, A.Y, Rochel, N, Guichard, G. | | Deposit date: | 2020-02-04 | | Release date: | 2021-02-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for alpha-Helix Mimicry and Inhibition of Protein-Protein Interactions with Oligourea Foldamers.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6X6B
 
 | |
1DFP
 
 | | FACTOR D INHIBITED BY DIISOPROPYL FLUOROPHOSPHATE | | Descriptor: | DIISOPROPYL PHOSPHONATE, FACTOR D | | Authors: | Cole, L.B, Chu, N, Kilpatrick, J.M, Volanakis, J.E, Narayana, S.V.L, Babu, Y.S. | | Deposit date: | 1997-02-18 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of diisopropyl fluorophosphate-inhibited factor D.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
6XZK
 
 | | Structure of zVDR LBD-Calcitriol in complex with chimera 13 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, ACETATE ION, GLU-ASN-ALA-UIA-URL-URY-URV-UZN-LYS, ... | | Authors: | Buratto, J, Belorusova, A.Y, Rochel, N, Guichard, G. | | Deposit date: | 2020-02-04 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for alpha-Helix Mimicry and Inhibition of Protein-Protein Interactions with Oligourea Foldamers.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6L2E
 
 | | Crystal structure of a cupin protein (tm1459, H52A mutant) in copper (Cu) substituted form | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, Cupin_2 domain-containing protein | | Authors: | Fujieda, N, Ichihashi, H, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-10-03 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.201 Å) | | Cite: | Cupin Variants as a Macromolecular Ligand Library for Stereoselective Michael Addition of Nitroalkanes.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
1DK1
 
 | | DETAILED VIEW OF A KEY ELEMENT OF THE RIBOSOME ASSEMBLY: CRYSTAL STRUCTURE OF THE S15-RRNA COMPLEX | | Descriptor: | 30S RIBOSOMAL PROTEIN S15, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Nikulin, A, Serganov, A, Ennifar, E, Tischenko, S, Nevskaya, N. | | Deposit date: | 1999-12-06 | | Release date: | 2000-04-02 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the S15-rRNA complex.
Nat.Struct.Biol., 7, 2000
|
|
6XB9
 
 | | Crystal structure of Azotobacter vinelandii 3-mercaptopropionic acid dioxygenase in complex with 3-hydroxypropionic acid | | Descriptor: | 3-HYDROXY-PROPANOIC ACID, CHLORIDE ION, Cysteine dioxygenase type I protein, ... | | Authors: | Kiser, P.D, Khadka, N, Shi, W, Pierce, B.S. | | Deposit date: | 2020-06-05 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of 3-mercaptopropionic acid dioxygenase with a substrate analog reveals bidentate substrate binding at the iron center.
J.Biol.Chem., 296, 2021
|
|
6X93
 
 | | Interleukin-10 signaling complex with IL-10RA and IL-10RB | | Descriptor: | Interleukin-10, Interleukin-10 receptor subunit alpha, Interleukin-10 receptor subunit beta | | Authors: | Saxton, R.A, Tsutsumi, N, Gati, C, Garcia, K.C. | | Deposit date: | 2020-06-02 | | Release date: | 2021-03-17 | | Last modified: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure-based decoupling of the pro- and anti-inflammatory functions of interleukin-10.
Science, 371, 2021
|
|
6L7X
 
 | | Crystal structure of Cet1 from Trypanosoma cruzi in complex with #951 ligand | | Descriptor: | 1,3-dimethyl-7-propyl-purine-2,6-dione, SULFATE ION, mRNA_triPase domain-containing protein | | Authors: | Kuwabara, N, Ho, K. | | Deposit date: | 2019-11-03 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structures of the RNA triphosphatase fromTrypanosoma cruziprovide insights into how it recognizes the 5'-end of the RNA substrate.
J.Biol.Chem., 295, 2020
|
|
1DB1
 
 | | CRYSTAL STRUCTURE OF THE NUCLEAR RECEPTOR FOR VITAMIN D COMPLEXED TO VITAMIN D | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, VITAMIN D NUCLEAR RECEPTOR | | Authors: | Rochel, N, Wurtz, J.M, Mitschler, A, Klaholz, B, Moras, D. | | Deposit date: | 1999-11-02 | | Release date: | 2000-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the nuclear receptor for vitamin D bound to its natural ligand.
Mol.Cell, 5, 2000
|
|
6L5V
 
 | | Carbonmonoxy human hemoglobin C in the R quaternary structure at 95 K: Light (2 min) | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Shibayama, N, Park, S.Y, Ohki, M, Sato-Tomita, A. | | Deposit date: | 2019-10-24 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Direct observation of ligand migration within human hemoglobin at work.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
