8UG4
 
 | |
6L4U
 
 | | Structure of the PSI-FCPI supercomplex from diatom | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'-yl acetate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Nagao, R, Kato, K, Miyazaki, N, Akita, F, Shen, J.R. | | Deposit date: | 2019-10-21 | | Release date: | 2020-05-20 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structure of photosystem I-light-harvesting supercomplex from a red-lineage diatom
Nat Commun, 2020
|
|
9IYH
 
 | | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter spp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.25 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHONOACETIC ACID, ... | | Authors: | Ahmad, N, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2024-07-30 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter spp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.25 A resolution.
To Be Published
|
|
8VPU
 
 | |
9GDK
 
 | | Jumonji domain-containing protein 1C with crystallization epitope mutations L2440Y:G2444H | | Descriptor: | Probable JmjC domain-containing histone demethylation protein 2C | | Authors: | Fairhead, M, Strain-Damerell, C, Ye, M, Mackinnon, S.R, Pinkas, D, MacLean, E.M, Koekemoer, L, Damerell, D, Krojer, T, Arrowsmith, C.H, Edwards, A, Bountra, C, Yue, W, Burgess-Brown, N, Marsden, B, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-08-05 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A fast, parallel method for efficiently exploring crystallization behaviour of large numbers of protein variants
To Be Published
|
|
6L4T
 
 | | Structure of the peripheral FCPI from diatom | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'-yl acetate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Nagao, R, Kato, K, Miyazaki, N, Akita, F, Shen, J.R. | | Deposit date: | 2019-10-21 | | Release date: | 2020-05-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure of photosystem I-light-harvesting supercomplex from a red-lineage diatom
Nat Commun, 2020
|
|
9IYG
 
 | |
5JX0
 
 | | Temperature sensitive D4 mutant L110F | | Descriptor: | CHLORIDE ION, GLYCEROL, Uracil-DNA glycosylase | | Authors: | Schormann, N, Chattopadhyay, D. | | Deposit date: | 2016-05-12 | | Release date: | 2017-02-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Poxvirus uracil-DNA glycosylase-An unusual member of the family I uracil-DNA glycosylases.
Protein Sci., 25, 2016
|
|
8UG6
 
 | |
5KER
 
 | | Deer mouse recombinant hemoglobin from high altitude species | | Descriptor: | Alpha-globin, Beta globin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Inoguchi, N, Natarajan, C, Storz, J.F, Moriyama, H. | | Deposit date: | 2016-06-10 | | Release date: | 2017-04-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Alteration of the alpha 1 beta 2/ alpha 2 beta 1 subunit interface contributes to the increased hemoglobin-oxygen affinity of high-altitude deer mice.
PLoS ONE, 12, 2017
|
|
8UG7
 
 | |
8UG8
 
 | |
8UGA
 
 | |
9GH3
 
 | | pleckstrin homology domain interacting protein with crystallization epitope mutations L1408N:R1409E | | Descriptor: | 1,2-ETHANEDIOL, PH-interacting protein | | Authors: | Fairhead, M, Strain-Damerell, C, Ye, M, Mackinnon, S.R, Pinkas, D, MacLean, E.M, Koekemoer, L, Damerell, D, Krojer, T, Arrowsmith, C.H, Edwards, A, Yue, W, Burgess-Brown, N, Marsden, B, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-08-14 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | A fast, parallel method for efficiently exploring crystallization behaviour of large numbers of protein variants
To Be Published
|
|
9F1B
 
 | | Mammalian ternary complex of a translating 80S ribosome, NAC and NatA/E | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Yudin, D, Scaiola, A, Ban, N. | | Deposit date: | 2024-04-18 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | NAC guides a ribosomal multienzyme complex for nascent protein processing
Nature, 2024
|
|
5ZQZ
 
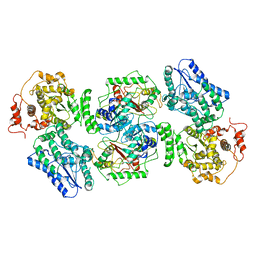 | | Structure of human mitochondrial trifunctional protein, tetramer | | Descriptor: | Trifunctional enzyme subunit alpha, mitochondrial, Trifunctional enzyme subunit beta | | Authors: | Liang, K, Li, N, Dai, J, Wang, X, Liu, P, Chen, X, Wang, C, Gao, N, Xiao, J. | | Deposit date: | 2018-04-20 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of human mitochondrial trifunctional protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6XYC
 
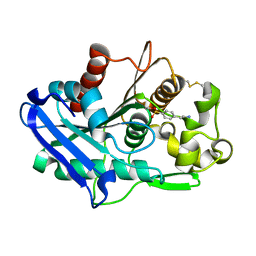 | | Truncated form of carbohydrate esterase from gut microbiota | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, Acetyl xylan esterase | | Authors: | Penttinen, L, Hakulinen, N, Master, E. | | Deposit date: | 2020-01-30 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Polysaccharide utilization loci-driven enzyme discovery reveals BD-FAE: a bifunctional feruloyl and acetyl xylan esterase active on complex natural xylans.
Biotechnol Biofuels, 14, 2021
|
|
6XXX
 
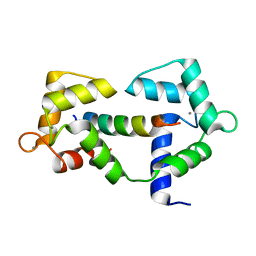 | | 1.25 Angstrom crystal structure of Ca/CaM A102V:RyR2 peptide complex | | Descriptor: | CALCIUM ION, Calmodulin-1, LYS-LYS-ALA-VAL-TRP-HIS-LYS-LEU-LEU-SER-LYS-GLN-ARG-LYS-ARG-ALA-VAL-VAL-ALA-CYS-PHE | | Authors: | Antonyuk, S, Helassa, N. | | Deposit date: | 2020-01-28 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | CPVT-associated calmodulin variants N53I and A102V dysregulate Ca2+ signalling via different mechanisms.
J.Cell.Sci., 135, 2022
|
|
121D
 
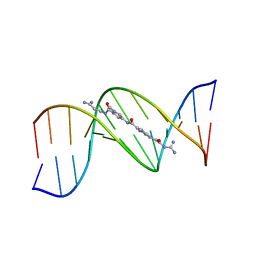 | | MOLECULAR STRUCTURE OF THE A-TRACT DNA DODECAMER D(CGCAAATTTGCG) COMPLEXED WITH THE MINOR GROOVE BINDING DRUG NETROPSIN | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*G)-3'), NETROPSIN | | Authors: | Tabernero, L, Verdaguer, N, Coll, M, Fita, I, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Aymami, J. | | Deposit date: | 1993-04-14 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular structure of the A-tract DNA dodecamer d(CGCAAATTTGCG) complexed with the minor groove binding drug netropsin.
Biochemistry, 32, 1993
|
|
1AHM
 
 | | DER F 2, THE MAJOR MITE ALLERGEN FROM DERMATOPHAGOIDES FARINAE, NMR, 10 STRUCTURES | | Descriptor: | DER F 2 | | Authors: | Ichikawa, S, Hatanaka, H, Yuuki, T, Iwamoto, N, Ogura, K, Okumura, Y, Inagaki, F. | | Deposit date: | 1997-04-07 | | Release date: | 1998-04-08 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Der f 2, the major mite allergen for atopic diseases.
J.Biol.Chem., 273, 1998
|
|
6ZOP
 
 | | Structure of the cysteine-rich domain of PiggyMac, a domesticated PiggyBac transposase involved in programmed genome rearrangements | | Descriptor: | DDE_Tnp_1_7 domain-containing protein, ZINC ION | | Authors: | Bessa, L, Guerineau, M, Moriau, S, Lescop, E, Bontems, F, Mathy, N, Guittet, E, Bischerour, J, Betermier, M, Morellet, N. | | Deposit date: | 2020-07-07 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | SOLUTION NMR | | Cite: | The unusual structure of the PiggyMac cysteine-rich domain reveals zinc finger diversity in PiggyBac-related transposases.
Mob DNA, 12, 2021
|
|
5W1V
 
 | | Structure of the HLA-E-VMAPRTLIL/GF4 TCR complex | | Descriptor: | Beta-2-microglobulin, GF4 T cell receptor alpha chain, GF4 T cell receptor beta chain, ... | | Authors: | Gras, S, Walpole, N, Farenc, C, Rossjohn, J. | | Deposit date: | 2017-06-04 | | Release date: | 2017-10-04 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | A conserved energetic footprint underpins recognition of human leukocyte antigen-E by two distinct alpha beta T cell receptors.
J. Biol. Chem., 292, 2017
|
|
5K31
 
 | | Crystal structure of Human fibrillar procollagen type I C-propeptide Homo-trimer | | Descriptor: | CALCIUM ION, CHLORIDE ION, Collagen alpha-1(I) chain, ... | | Authors: | Sharma, U, Hulmes, D.J.S, Aghajari, N. | | Deposit date: | 2016-05-19 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of homo- and heterotrimerization of collagen I.
Nat Commun, 8, 2017
|
|
5VL4
 
 | | Accidental minimum contact crystal lattice formed by a redesigned protein oligomer | | Descriptor: | T33-53H-B | | Authors: | Cannon, K.A, Cascio, D, Sawaya, M.R, Park, R, Boyken, S, King, N, Yeates, T. | | Deposit date: | 2017-04-24 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Design and structure of two new protein cages illustrate successes and ongoing challenges in protein engineering.
Protein Sci., 29, 2020
|
|
1A1Q
 
 | | HEPATITIS C VIRUS NS3 PROTEINASE | | Descriptor: | NS3 PROTEINASE, ZINC ION | | Authors: | Love, R.A, Parge, H.E, Wickersham, J.A, Hostomsky, Z, Habuka, N, Moomaw, E.W, Adachi, T, Hostomska, Z. | | Deposit date: | 1997-12-12 | | Release date: | 1998-03-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of hepatitis C virus NS3 proteinase reveals a trypsin-like fold and a structural zinc binding site.
Cell(Cambridge,Mass.), 87, 1996
|
|
