5KER
 
 | | Deer mouse recombinant hemoglobin from high altitude species | | Descriptor: | Alpha-globin, Beta globin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Inoguchi, N, Natarajan, C, Storz, J.F, Moriyama, H. | | Deposit date: | 2016-06-10 | | Release date: | 2017-04-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Alteration of the alpha 1 beta 2/ alpha 2 beta 1 subunit interface contributes to the increased hemoglobin-oxygen affinity of high-altitude deer mice.
PLoS ONE, 12, 2017
|
|
6LTL
 
 | | The dimeric structure of G80A myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagao, S, Suda, A, Kobayashi, H, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2020-01-22 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Thermodynamic Control of Domain Swapping by Modulating the Helical Propensity in the Hinge Region of Myoglobin.
Chem Asian J, 15, 2020
|
|
6M0X
 
 | | Crystal structure of Streptococcus thermophilus Cas9 in complex with AGGA PAM | | Descriptor: | BARIUM ION, CRISPR-associated endonuclease Cas9 1, DNA (28-MER), ... | | Authors: | Zhang, Y, Zhang, H, Xu, X, Wang, Y, Chen, W, Wang, Y, Wu, Z, Tang, N, Wang, Y, Zhao, S, Gan, J, Ji, Q. | | Deposit date: | 2020-02-23 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.561 Å) | | Cite: | Catalytic-state structure and engineering of Streptococcus thermophilus Cas9
Nat Catal, 2020
|
|
5JX0
 
 | | Temperature sensitive D4 mutant L110F | | Descriptor: | CHLORIDE ION, GLYCEROL, Uracil-DNA glycosylase | | Authors: | Schormann, N, Chattopadhyay, D. | | Deposit date: | 2016-05-12 | | Release date: | 2017-02-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Poxvirus uracil-DNA glycosylase-An unusual member of the family I uracil-DNA glycosylases.
Protein Sci., 25, 2016
|
|
1ISS
 
 | | Crystal Structure of Metabotropic Glutamate Receptor Subtype 1 Complexed with an antagonist | | Descriptor: | (S)-(ALPHA)-METHYL-4-CARBOXYPHENYLGLYCINE, Metabotropic Glutamate Receptor subtype 1 | | Authors: | Tsuchiya, D, Kunishima, N, Kamiya, N, Jingami, H, Morikawa, K. | | Deposit date: | 2001-12-21 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural views of the ligand-binding cores of a metabotropic glutamate receptor complexed with an antagonist and both glutamate and Gd3+.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
5K31
 
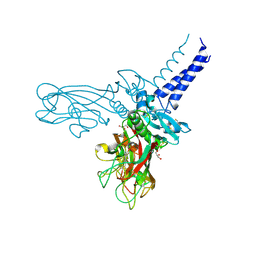 | | Crystal structure of Human fibrillar procollagen type I C-propeptide Homo-trimer | | Descriptor: | CALCIUM ION, CHLORIDE ION, Collagen alpha-1(I) chain, ... | | Authors: | Sharma, U, Hulmes, D.J.S, Aghajari, N. | | Deposit date: | 2016-05-19 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of homo- and heterotrimerization of collagen I.
Nat Commun, 8, 2017
|
|
6YNQ
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2-Methyl-1-tetralone. | | Descriptor: | (2~{S})-2-methyl-3,4-dihydro-2~{H}-naphthalen-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7ATK
 
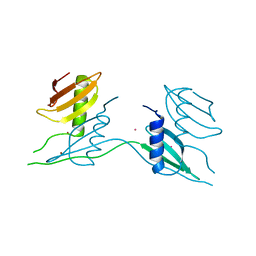 | | Crystal structure of UipA in complex with Uranium | | Descriptor: | URANIUM ATOM, UipA, ZINC ION | | Authors: | Bremond, N, Gallois, N, Legrand, P, Chapon, V, Arnoux, P. | | Deposit date: | 2020-10-30 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.855 Å) | | Cite: | Discovery and characterization of UipA, a uranium- and iron-binding PepSY protein involved in uranium tolerance by soil bacteria.
Isme J, 16, 2022
|
|
7ATH
 
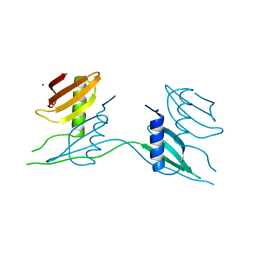 | | Crystal structure of UipA | | Descriptor: | UipA, ZINC ION | | Authors: | Bremond, N, Gallois, N, Legrand, P, Chapon, V, Arnoux, P. | | Deposit date: | 2020-10-30 | | Release date: | 2021-10-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.343 Å) | | Cite: | Discovery and characterization of UipA, a uranium- and iron-binding PepSY protein involved in uranium tolerance by soil bacteria.
Isme J, 16, 2022
|
|
1O3X
 
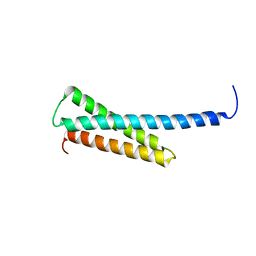 | | Crystal structure of human GGA1 GAT domain | | Descriptor: | ADP-ribosylation factor binding protein GGA1 | | Authors: | Shiba, T, Kawasaki, M, Takatsu, H, Nogi, T, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2003-05-08 | | Release date: | 2003-05-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Mechanism of Membrane Recruitment of Gga by Arf in Lysosomal Protein Transport
Nat.Struct.Biol., 10, 2003
|
|
7N82
 
 | | NMR Solution structure of Se0862 | | Descriptor: | Biofilm-related protein | | Authors: | Zhang, N, LiWang, A.L. | | Deposit date: | 2021-06-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Assessment of prediction methods for protein structures determined by NMR in CASP14: Impact of AlphaFold2.
Proteins, 89, 2021
|
|
6NEP
 
 | | Structure of human PACRG-MEIG1 complex | | Descriptor: | CHLORIDE ION, Meiosis expressed gene 1 protein homolog, PHOSPHATE ION, ... | | Authors: | Khan, N, Croteau, N, Pelletier, D, Veyron, S, Trempe, J.F. | | Deposit date: | 2018-12-18 | | Release date: | 2019-10-23 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.097625 Å) | | Cite: | Crystal structure of human PACRG in complex with MEIG1
Biorxiv, 2019
|
|
1O3Y
 
 | | Crystal structure of mouse ARF1 (delta17-Q71L), GTP form | | Descriptor: | ADP-ribosylation factor 1, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Shiba, T, Kawasaki, M, Takatsu, H, Nogi, T, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2003-05-08 | | Release date: | 2003-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular mechanism of membrane recruitment of GGA by ARF in lysosomal protein transport
Nat.Struct.Biol., 10, 2003
|
|
4UBQ
 
 | | Crystal Structure of IMP-2 Metallo-beta-Lactamase from Acinetobacter spp. | | Descriptor: | ACETATE ION, Beta-lactamase, ZINC ION | | Authors: | Yamaguchi, Y, Matsueda, S, Matsunaga, K, Takashio, N, Toma-Fukai, S, Yamagata, Y, Shibata, N, Wachino, J, Shibayama, K, Arakawa, Y, Kurosaki, H. | | Deposit date: | 2014-08-13 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of IMP-2 metallo-beta-lactamase from Acinetobacter spp.: comparison of active-site loop structures between IMP-1 and IMP-2.
Biol.Pharm.Bull., 38, 2015
|
|
6YVF
 
 | | Structure of SARS-CoV-2 Main Protease bound to AZD6482. | | Descriptor: | 2-[[(1R)-1-(7-methyl-2-morpholin-4-yl-4-oxidanylidene-pyrido[1,2-a]pyrimidin-9-yl)ethyl]amino]benzoic acid, 3C-like proteinase, CALCIUM ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
4V1A
 
 | | Structure of the large subunit of the mammalian mitoribosome, part 2 of 2 | | Descriptor: | MITORIBOSOMAL PROTEIN ML37, MRPL37, MITORIBOSOMAL PROTEIN ML38, ... | | Authors: | Greber, B.J, Boehringer, D, Leibundgut, M, Bieri, P, Leitner, A, Schmitz, N, Aebersold, R, Ban, N. | | Deposit date: | 2014-09-25 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The Complete Structure of the Large Subunit of the Mammalian Mitochondrial Ribosome
Nature, 515, 2014
|
|
1ON7
 
 | |
5EC5
 
 | | Crystal structure of lysenin pore | | Descriptor: | Lysenin, MERCURIBENZOIC ACID, MERCURY (II) ION | | Authors: | Podobnik, M, Savory, P, Rojko, N, Kisovec, M, Bruce, M, Jayasinghe, L, Anderluh, G. | | Deposit date: | 2015-10-20 | | Release date: | 2016-05-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of an invertebrate cytolysin pore reveals unique properties and mechanism of assembly.
Nat Commun, 7, 2016
|
|
4U8H
 
 | | Crystal Structure of Mammalian Period-Cryptochrome Complex | | Descriptor: | Cryptochrome-2, Period circadian protein homolog 2, ZINC ION | | Authors: | Nangle, S.N, Rosensweig, C, Koike, N, Tei, H, Takahashi, J.S, Green, C.B, Zheng, N. | | Deposit date: | 2014-08-03 | | Release date: | 2014-10-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Molecular assembly of the period-cryptochrome circadian transcriptional repressor complex.
Elife, 3, 2014
|
|
4U46
 
 | | Crystal structure of an avidin mutant | | Descriptor: | Avidin, CHLORIDE ION | | Authors: | Agrawal, N, Lehtonen, S, Kahkonen, N, Riihimaki, T, Hytonen, V.P, Kulomaa, M.S, Johnson, M.S, Airenne, T.T. | | Deposit date: | 2014-07-23 | | Release date: | 2015-08-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Artificial Avidin-Based Receptors for a Panel of Small Molecules.
Acs Chem.Biol., 11, 2016
|
|
6NDU
 
 | | Structure of human PACRG-MEIG1 complex | | Descriptor: | CHLORIDE ION, Meiosis expressed gene 1 protein homolog, PHOSPHATE ION, ... | | Authors: | Khan, N, Croteau, N, Pelletier, D, Veyron, S, Trempe, J.F. | | Deposit date: | 2018-12-14 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human PACRG in complex with MEIG1
Biorxiv, 2019
|
|
4V11
 
 | | Structure of Synaptotagmin-1 with SV2A peptide phosphorylated at Thr84 | | Descriptor: | CALCIUM ION, GLYCEROL, SYNAPTIC VESICLE GLYCOPROTEIN 2A, ... | | Authors: | Zhang, N, Gordon, S.L, Fritsch, M.J, Esoof, N, Campbell, D, Gourlay, R, Velupillai, S, Macartney, T, Peggie, M, vanAalten, D.M.F, Cousin, M.A, Alessi, D.R. | | Deposit date: | 2014-09-22 | | Release date: | 2015-02-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Phosphorylation of Synaptic Vesicle Protein 2A at Thr84 by Casein Kinase 1 Family Kinases Controls the Specific Retrieval of Synaptotagmin-1.
J.Neurosci., 35, 2015
|
|
5J0N
 
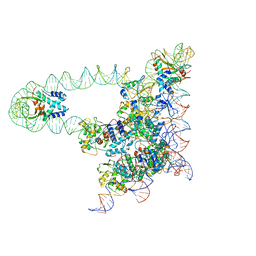 | | Lambda excision HJ intermediate | | Descriptor: | Excisionase, Integrase, Integration host factor subunit alpha, ... | | Authors: | Van Duyne, G, Grigorieff, N, Landy, A. | | Deposit date: | 2016-03-28 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Structure of a Holliday junction complex reveals mechanisms governing a highly regulated DNA transaction.
Elife, 5, 2016
|
|
1NHU
 
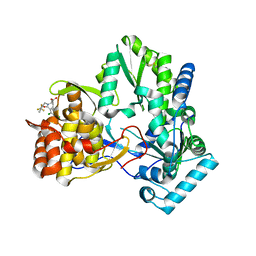 | | Hepatitis C virus RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | (2S)-2-[(2,4-DICHLORO-BENZOYL)-(3-TRIFLUOROMETHYL-BENZYL)-AMINO]-3-PHENYL-PROPIONIC ACID, HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE | | Authors: | Wang, M, Ng, K.K.S, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bedard, J, Morin, N, Nguyen-Ba, N, Alaoui-Ismaili, M.H, Bethell, R.C, James, M.N.G. | | Deposit date: | 2002-12-19 | | Release date: | 2003-03-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Non-Nucleoside Analogue Inhibitors Bind to an Allosteric Site on
HCV NS5B Polymerase: Crystal Structures and Mechanism of Inhibition
J.Biol.Chem., 278, 2003
|
|
5LS6
 
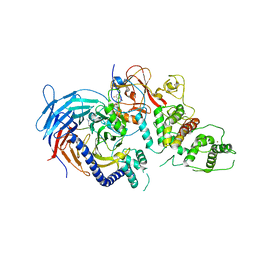 | | Structure of Human Polycomb Repressive Complex 2 (PRC2) with inhibitor | | Descriptor: | 1-[(1~{R})-1-[1-[2,2-bis(fluoranyl)propyl]piperidin-4-yl]ethyl]-~{N}-[(4-methoxy-6-methyl-2-oxidanylidene-3~{H}-pyridin-3-yl)methyl]-2-methyl-indole-3-carboxamide, Histone-lysine N-methyltransferase EZH2,Histone-lysine N-methyltransferase EZH2,Histone-lysine N-methyltransferase EZH2, Jarid2 K116me3, ... | | Authors: | Zhang, Y, Justin, N, Chen, S, Wilson, J, Gamblin, S. | | Deposit date: | 2016-08-22 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Identification of (R)-N-((4-Methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-2-methyl-1-(1-(1-(2,2,2-trifluoroethyl)piperidin-4-yl)ethyl)-1H-indole-3-carboxamide (CPI-1205), a Potent and Selective Inhibitor of Histone Methyltransferase EZH2, Suitable for Phase I Clinical Trials for B-Cell Lymphomas.
J. Med. Chem., 59, 2016
|
|
