6U20
 
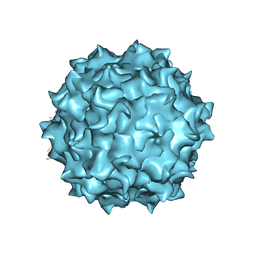 | | AAV8 human HEK293-produced, empty capsid | | Descriptor: | Capsid protein | | Authors: | Paulk, N.K, Poweleit, N. | | Deposit date: | 2019-08-18 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Methods Matter: Standard Production Platforms for Recombinant AAV Produce Chemically and Functionally Distinct Vectors.
Mol Ther Methods Clin Dev, 18, 2020
|
|
6U2Y
 
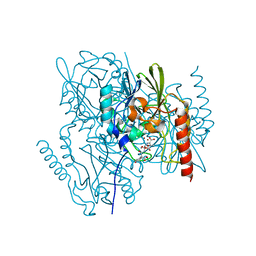 | | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the hydrolyzed moxalactam and two Ni ions | | Descriptor: | (2R)-2-((R)-CARBOXY{[CARBOXY(4-HYDROXYPHENYL)ACETYL]AMINO}METHOXYMETHYL)-5-METHYLENE-5,6-DIHYDRO-2H-1,3-OXAZINE-4-CARBO XYLIC ACID, 1,2-ETHANEDIOL, NICKEL (II) ION, ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-20 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the hydrolyzed moxalactam and two Ni ions
To Be Published
|
|
6TVG
 
 | |
6GV9
 
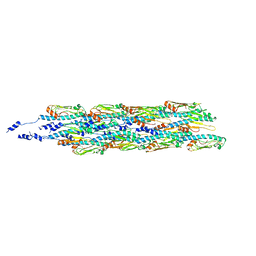 | | Structure of the type IV pilus from enterohemorrhagic Escherichia coli (EHEC) | | Descriptor: | Prepilin peptidase-dependent protein D | | Authors: | Bardiaux, B, Amorim, G.C, Luna-Rico, A, Zheng, W, Guilvout, I, Jollivet, C, Nilges, M, Egelman, E, Francetic, O, Izadi-Pruneyre, N. | | Deposit date: | 2018-06-20 | | Release date: | 2019-05-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structure and Assembly of the Enterohemorrhagic Escherichia coli Type 4 Pilus.
Structure, 27, 2019
|
|
6GVL
 
 | | Second pair of Fibronectin type III domains of integrin beta4 bound to the bullous pemphigoid antigen BP230 (BPAG1e) | | Descriptor: | Dystonin, Integrin beta-4 | | Authors: | Manso, J.A, Gomez-Hernandez, M, Alonso-Garcia, N, de Pereda, J.M. | | Deposit date: | 2018-06-21 | | Release date: | 2019-03-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Integrin alpha 6 beta 4 Recognition of a Linear Motif of Bullous Pemphigoid Antigen BP230 Controls Its Recruitment to Hemidesmosomes.
Structure, 27, 2019
|
|
6GP2
 
 | | Ribonucleotide Reductase class Ie R2 from Mesoplasma florum, DOPA-active form | | Descriptor: | CALCIUM ION, Ribonucleoside-diphosphate reductase beta chain | | Authors: | Srinivas, V, Lebrette, H, Lundin, D, Kutin, Y, Sahlin, M, Lerche, M, Enrich, J, Branca, R.M.M, Cox, N, Sjoberg, B.M, Hogbom, M. | | Deposit date: | 2018-06-05 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Metal-free ribonucleotide reduction powered by a DOPA radical in Mycoplasma pathogens.
Nature, 563, 2018
|
|
6DSY
 
 | | Bst DNA polymerase I post-chemistry (n+1) structure | | Descriptor: | DNA (5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*T)-3'), DNA (5'-D(P*GP*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3'), DNA polymerase I, ... | | Authors: | Chim, N, Jackson, L.N, Chaput, J.C. | | Deposit date: | 2018-06-14 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structures of DNA polymerase I capture novel intermediates in the DNA synthesis pathway.
Elife, 7, 2018
|
|
6PY9
 
 | | Crystal structure of red kidney bean purple acid phosphatase in complex with adenosine diphosphate metavanadate | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADP METAVANADATE, ... | | Authors: | Feder, D, Schenk, G, Guddat, L.W, McGeary, R.P, Mitic, N, Furtado, A, Schulz, B.L, Henry, R.J, Schmidt, S. | | Deposit date: | 2019-07-29 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural elements that modulate the substrate specificity of plant purple acid phosphatases: Avenues for improved phosphorus acquisition in crops.
Plant Sci., 294, 2020
|
|
6GFI
 
 | | Structure of Human Mesotrypsin in complex with APPI variant T11V/M17R/I18F/F34V | | Descriptor: | 1,2-ETHANEDIOL, Amyloid-beta A4 protein, PRSS3 protein | | Authors: | Shahar, A, Cohen, I, Radisky, E, Papo, N, Naftaly, S. | | Deposit date: | 2018-04-30 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mapping protein selectivity landscapes using multi-target selective screening and next-generation sequencing of combinatorial libraries.
Nat Commun, 9, 2018
|
|
6TZV
 
 | | Crystal Structure of the carboxyltransferase subunit of ACC (AccD6) in complex with inhibitor Phenyl-Cyclodiaone from Mycobacterium tuberculosis | | Descriptor: | 4'-[(6-chloro-1,3-benzothiazol-2-yl)oxy]-6-hydroxy-4,4-dimethyl-4,5-dihydro[1,1'-biphenyl]-2(3H)-one, Probable propionyl-CoA carboxylase beta chain 6 | | Authors: | Reddy, M.C.M, Zhou, N, Sacchettini, J, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2019-08-13 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.388 Å) | | Cite: | Novel herbicidal derivatives that inhibit carboxyltransferase subunit of the acetyl-CoA carboxylase in Mycobacterium tuberculosis
To Be Published
|
|
6U10
 
 | | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the inhibitor captopril | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, L-CAPTOPRIL, ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-15 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the inhibitor captopril.
To Be Published
|
|
6GHD
 
 | | Structural analysis of the ternary complex between lamin A/C, BAF and emerin identifies an interface disrupted in autosomal recessive progeroid diseases | | Descriptor: | 1,2-ETHANEDIOL, Barrier-to-autointegration factor, Emerin, ... | | Authors: | Samson, C, Petitalot, A, Celli, F, Herrada, I, Ropars, V, Ledu, M.H, Nhiri, N, Arteni, A.A, Buendia, B, ZinnJustin, S. | | Deposit date: | 2018-05-07 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of the ternary complex between lamin A/C, BAF and emerin identifies an interface disrupted in autosomal recessive progeroid diseases.
Nucleic Acids Res., 46, 2018
|
|
5CQ4
 
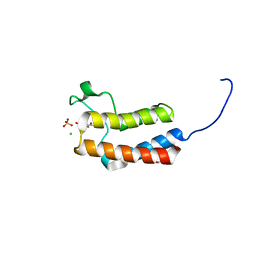 | | Crystal structure of the bromodomain of bromodomain adjacent to zinc finger domain protein 2B (BAZ2B) in complex with 3'-Hydroxyacetophenone (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-hydroxyphenyl)ethanone, Bromodomain adjacent to zinc finger domain protein 2B, ... | | Authors: | Bradley, A, Pearce, N, Krojer, T, Ng, J, Talon, R, Vollmar, M, Jose, B, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-21 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.782 Å) | | Cite: | Crystal structure of the second bromodomain of bromodomain adjancent to zinc finger domain protein 2B (BAZ2B) in complex with 3'-Hydroxyacetophenone (SGC - Diamond I04-1 fragment screening)
To be published
|
|
6GPD
 
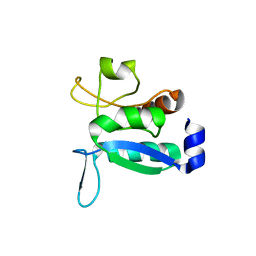 | | Crystal structure of the ligand-free form of domain 1 from TmArgBP | | Descriptor: | Amino acid ABC transporter, periplasmic amino acid-binding protein,Amino acid ABC transporter, periplasmic amino acid-binding protein | | Authors: | Smaldone, G, Balasco, N, Ruggiero, A, Berisio, R, Vitagliano, L. | | Deposit date: | 2018-06-05 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Domain communication in Thermotoga maritima Arginine Binding Protein unraveled through protein dissection.
Int. J. Biol. Macromol., 119, 2018
|
|
6Q2K
 
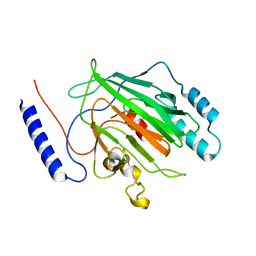 | |
6U2V
 
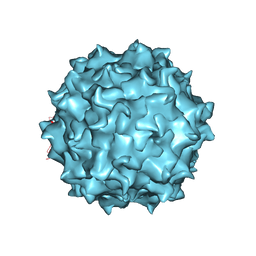 | | AAV8 Baculovirus-Sf9 produced, full capsid | | Descriptor: | Capsid protein | | Authors: | Paulk, N.K, Poweleit, N. | | Deposit date: | 2019-08-20 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Methods Matter: Standard Production Platforms for Recombinant AAV Produce Chemically and Functionally Distinct Vectors.
Mol Ther Methods Clin Dev, 18, 2020
|
|
1R1B
 
 | | EPRS SECOND REPEATED ELEMENT, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | TRNA SYNTHETASE | | Authors: | Cahuzac, B, Berthonneau, E, Birlirakis, N, Mirande, M, Guittet, E. | | Deposit date: | 1998-12-15 | | Release date: | 1999-12-15 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A recurrent RNA-binding domain is appended to eukaryotic aminoacyl-tRNA synthetases.
EMBO J., 19, 2000
|
|
6Q64
 
 | | BT1044SeMet E190Q | | Descriptor: | Endoglycosidase | | Authors: | Basle, A, Paterson, N, Crouch, L, Bolam, D. | | Deposit date: | 2018-12-10 | | Release date: | 2019-05-08 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Complex N-glycan breakdown by gut Bacteroides involves an extensive enzymatic apparatus encoded by multiple co-regulated genetic loci.
Nat Microbiol, 4, 2019
|
|
1R53
 
 | | Crystal structure of the bifunctional chorismate synthase from Saccharomyces cerevisiae | | Descriptor: | Chorismate synthase | | Authors: | Quevillon-Cheruel, S, Leulliot, N, Meyer, P, Graille, M, Bremang, M, Blondeau, K, Sorel, I, Poupon, A, Janin, J, van Tilbeurgh, H. | | Deposit date: | 2003-10-09 | | Release date: | 2003-12-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the bifunctional chorismate synthase from Saccharomyces cerevisiae
J.Biol.Chem., 279, 2004
|
|
1R7C
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Minimized average structure, Sample in 50% tfe) | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
1R7F
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Ensemble of 43 structures. Sample in 100mM SDS) | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
6W5V
 
 | | NPC1-NPC2 complex structure at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Yan, N, Qian, H.W, Wu, X.L. | | Deposit date: | 2020-03-13 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural Basis of Low-pH-Dependent Lysosomal Cholesterol Egress by NPC1 and NPC2.
Cell, 182, 2020
|
|
6Q70
 
 | | Crystal structure of the alanine racemase Bsu17640 from Bacillus subtilis in the presence of HEPES | | Descriptor: | Alanine racemase 2, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bernardo-Garcia, N, Gago, F, Hermoso, J.A. | | Deposit date: | 2018-12-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Cold-induced aldimine bond cleavage by Tris in Bacillus subtilis alanine racemase.
Org.Biomol.Chem., 17, 2019
|
|
1R20
 
 | | Crystal structure of the ligand-binding domains of the heterodimer EcR/USP bound to the synthetic agonist BYI06830 | | Descriptor: | ECDYSONE RECEPTOR, L-ALPHA-PHOSPHATIDYL-BETA-OLEOYL-GAMMA-PALMITOYL-PHOSPHATIDYLETHANOLAMINE, N-(TERT-BUTYL)-3,5-DIMETHYL-N'-[(5-METHYL-2,3-DIHYDRO-1,4-BENZODIOXIN-6-YL)CARBONYL]BENZOHYDRAZIDE, ... | | Authors: | Billas, I.M.L, Iwema, T, Garnier, J.M, Mitschler, A, Rochel, N, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-09-25 | | Release date: | 2003-11-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural adaptability in the ligand-binding pocket of the ecdysone hormone receptor.
Nature, 426, 2003
|
|
6GIM
 
 | | Structure of the DNA duplex d(AAATTT)2 with [N-(3-chloro-4-((4,5-dihydro-1H-imidazol-2-yl)amino)phenyl)-4-((4,5-dihydro-1H-imidazol-2- yl)amino)benzamide] - (drug JNI18) | | Descriptor: | DNA (5'-D(*AP*AP*AP*TP*TP*T)-3'), MAGNESIUM ION, [4-[(3-chloranyl-4-imidazolidin-2-ylideneazaniumyl-phenyl)carbamoyl]phenyl]-imidazolidin-2-ylidene-azanium | | Authors: | Millan, C.R, Dardonvile, C, de Koning, H.P, Saperas, N, Campos, J.L. | | Deposit date: | 2018-05-14 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Functional and structural analysis of AT-specific minor groove binders that disrupt DNA-protein interactions and cause disintegration of the Trypanosoma brucei kinetoplast.
Nucleic Acids Res., 45, 2017
|
|
