7C94
 
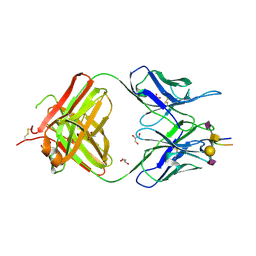 | | Crystal structure of the anti-human podoplanin antibody Fab fragment complex with glycopeptide | | Descriptor: | GLYCEROL, Heavy chain of Fab fragment, Light chain of Fab fragment, ... | | Authors: | Suzuki, K, Nakamura, S, Ogasawara, S, Naruchi, K, Shimabukuro, J, Tukahara, N, Kaneko, M.K, Kato, Y, Murata, T. | | Deposit date: | 2020-06-04 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal structure of an anti-podoplanin antibody bound to a disialylated O-linked glycopeptide.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
7CBF
 
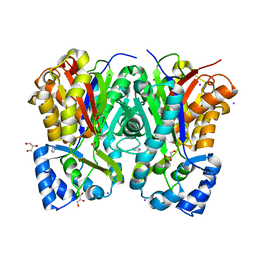 | | Crystal structure of benzophenone synthase from Garcinia mangostana L. pericarps reveals basis for substrate specificity and catalysis | | Descriptor: | 2,4,6-trihydroxybenzophenone synthase, GLYCEROL, IMIDAZOLE, ... | | Authors: | Songsiriritthigul, C, Nualkaew, N, Chen, C.-J. | | Deposit date: | 2020-06-12 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Crystal structure of benzophenone synthase from Garcinia mangostana L. pericarps reveals basis for substrate specificity and catalysis.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6CQI
 
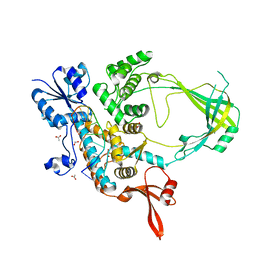 | | 2.42A Crystal structure of Mycobacterium tuberculosis Topoisomerase I in complex with an oligonucleotide MTS2-11 | | Descriptor: | ACETATE ION, DNA (5'-D(P*TP*TP*CP*CP*GP*CP*TP*TP*GP*A)-3'), DNA topoisomerase 1, ... | | Authors: | Cao, N, Thirunavukkarasu, A, Tan, K, Tse-Dinh, Y.-C. | | Deposit date: | 2018-03-15 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Investigating mycobacterial topoisomerase I mechanism from the analysis of metal and DNA substrate interactions at the active site.
Nucleic Acids Res., 46, 2018
|
|
6FE8
 
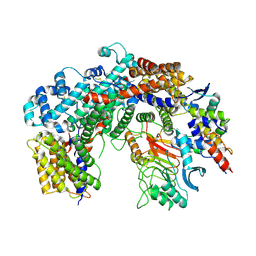 | | Cryo-EM structure of the core Centromere Binding Factor 3 complex | | Descriptor: | Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, Suppressor of kinetochore protein 1 | | Authors: | Zhang, W.J, Lukoynova, N, Miah, S, Vaughan, C.K. | | Deposit date: | 2017-12-30 | | Release date: | 2018-08-01 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Insights into Centromere DNA Bending Revealed by the Cryo-EM Structure of the Core Centromere Binding Factor 3 with Ndc10.
Cell Rep, 24, 2018
|
|
1QKF
 
 | | SOLUTION STRUCTURE OF THE RIBOSOMAL PROTEIN S19 FROM THERMUS THERMOPHILUS | | Descriptor: | 30S RIBOSOMAL PROTEIN S19 | | Authors: | Helgstrand, M, Rak, A.V, Allard, P, Davydova, N, Garber, M.B, Hard, T. | | Deposit date: | 1999-07-19 | | Release date: | 1999-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ribosomal protein S19 from Thermus thermophilus.
J. Mol. Biol., 292, 1999
|
|
6FFW
 
 | | Phosphotriesterase PTE_A53_5 | | Descriptor: | (4~{S},6~{R})-2,2,6-trimethyl-1,3-dioxan-4-ol, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Dym, O, Aggarwal, N, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Leader, H, Ashani, Y, Goldsmith, M, Greisen, P, Tawfik, D, Sussman, L.J. | | Deposit date: | 2018-01-09 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.495 Å) | | Cite: | Phosphotriesterase
PTE_A53_5
To Be Published
|
|
6CM2
 
 | | SAMHD1 HD domain bound to decitabine triphosphate | | Descriptor: | 6-amino-3-{2-deoxy-5-O-[(R)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-erythro-pentofuranosyl}-3,4-dihydro-1,3,5-triazin-2(1H)-one, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Oellerich, T, Schneider, C, Thomas, D, Knecht, K.M, Buzovetsky, O, Kaderali, L, Schliemann, C, Bohnenberger, H, Angenendt, L, Hartmann, W, Wardelmann, E, Rothenburger, T, Mohr, S, Scheich, S, Comoglio, F, Wilke, A, Strobel, P, Serve, H, Michaelis, M, Ferreiros, N, Geisslinger, G, Xiong, Y, Keppler, O.T, Cinatl, J. | | Deposit date: | 2018-03-02 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Selective inactivation of hypomethylating agents by SAMHD1 provides a rationale for therapeutic stratification in AML.
Nat Commun, 10, 2019
|
|
7ZEK
 
 | |
6CNA
 
 | | GluN1-GluN2B NMDA receptors with exon 5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Furukawa, H, Grant, T, Grigorieff, N. | | Deposit date: | 2018-03-07 | | Release date: | 2018-10-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural Mechanism of Functional Modulation by Gene Splicing in NMDA Receptors.
Neuron, 98, 2018
|
|
6FHO
 
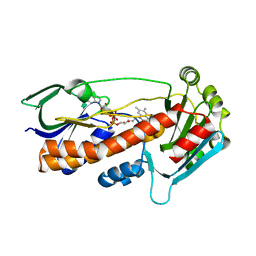 | | Crystal structure of pqsL, a probable FAD-dependent monooxygenase from Pseudomonas aeruginosa - new refinement | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Probable FAD-dependent monooxygenase | | Authors: | Belviso, B.D, Drees, S.L, Ernst, S, Jagmann, N, Hennecke, U, Fetzner, S. | | Deposit date: | 2018-01-15 | | Release date: | 2018-04-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | PqsL uses reduced flavin to produce 2-hydroxylaminobenzoylacetate, a preferred PqsBC substrate in alkyl quinolone biosynthesis inPseudomonas aeruginosa.
J. Biol. Chem., 293, 2018
|
|
6P1X
 
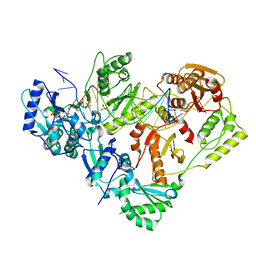 | | Structure of HIV-1 Reverse Transcriptase (RT) in complex with dsDNA and L-ddCTP | | Descriptor: | DNA Primer 20-mer, DNA template 27-mer, MAGNESIUM ION, ... | | Authors: | Bertoletti, N, Anderson, K.S. | | Deposit date: | 2019-05-20 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | Structural insights into the recognition of nucleoside reverse transcriptase inhibitors by HIV-1 reverse transcriptase: First crystal structures with reverse transcriptase and the active triphosphate forms of lamivudine and emtricitabine.
Protein Sci., 28, 2019
|
|
5CLT
 
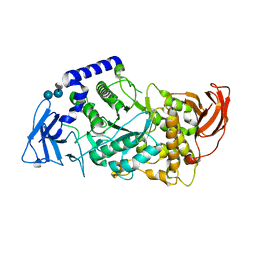 | | Crystal structure of human glycogen branching enzyme (GBE1) in complex with acarbose | | Descriptor: | 1,4-alpha-glucan-branching enzyme, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Krojer, T, Froese, D.S, Goubin, S, Strain-Damerell, C, Mahajan, P, Burgess-Brown, N, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Yue, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-16 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of human glycogen branching enzyme (GBE1) in complex with acarbose
To be published
|
|
5CQA
 
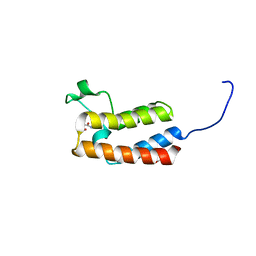 | | Crystal structure of the bromodomain of bromodomain adjacent to zinc finger domain protein 2B (BAZ2B) in complex with N-methyl-2,3-dihydrothieno[3,4-b][1,4]dioxine-5-carboxamide (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B, N-methyl-2,3-dihydrothieno[3,4-b][1,4]dioxine-5-carboxamide | | Authors: | Bradley, A, Pearce, N, Krojer, T, Ng, J, Talon, R, Vollmar, M, Jose, B, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-21 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of the second bromodomain of bromodomain adjancent to zinc finger domain protein 2B (BAZ2B) in complex with N-methyl-2,3-dihydrothieno[3,4-b][1,4]dioxine-5-carboxamide (SGC - Diamond I04-1 fragment screening)
To be published
|
|
6FKR
 
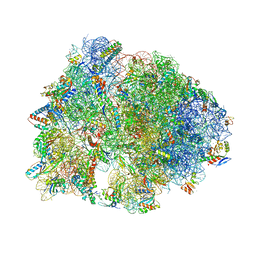 | | Crystal structure of the dolphin proline-rich antimicrobial peptide Tur1A bound to the Thermus thermophilus 70S ribosome | | Descriptor: | 16 ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mardirossian, M, Perebaskine, N, Benincasa, M, Gambato, S, Hofmann, S, Huter, P, Muller, C, Hilpert, K, Innis, C.A, Tossi, A, Wilson, D.N. | | Deposit date: | 2018-01-24 | | Release date: | 2018-03-28 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Dolphin Proline-Rich Antimicrobial Peptide Tur1A Inhibits Protein Synthesis by Targeting the Bacterial Ribosome.
Cell Chem Biol, 25, 2018
|
|
5CRY
 
 | | Structure of iron-saturated C-lobe of bovine lactoferrin at pH 6.8 indicates the softening of iron coordination | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BICARBONATE ION, FE (III) ION, ... | | Authors: | Singh, A, Rastogi, N, Singh, P.K, Tyagi, T.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-07-23 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure of iron-saturated C-lobe of bovine lactoferrin at pH 7.0 indicates the softening of iron coordination
To Be Published
|
|
1QVV
 
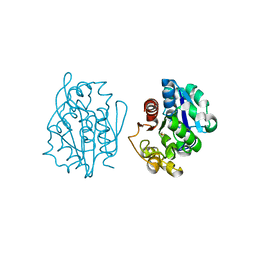 | | Crystal structure of the S. cerevisiae YDR533c protein | | Descriptor: | YDR533c protein | | Authors: | Graille, M, Leulliot, N, Quevillon-Cheruel, S, van Tilbeurgh, H. | | Deposit date: | 2003-08-29 | | Release date: | 2004-03-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the YDR533c S. cerevisiae protein, a class II member of the Hsp31 family
STRUCTURE, 12, 2004
|
|
6FLK
 
 | |
5CQ6
 
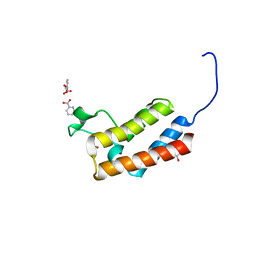 | | Crystal structure of the bromodomain of bromodomain adjacent to zinc finger domain protein 2B (BAZ2B) in complex with 2,6-Pyridinedicarboxylic acid (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B, PYRIDINE-2,6-DICARBOXYLIC ACID | | Authors: | Bradley, A, Pearce, N, Krojer, T, Ng, J, Talon, R, Vollmar, M, Jose, B, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-21 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of the second bromodomain of bromodomain adjancent to zinc finger domain protein 2B (BAZ2B) in complex with 2,6-Pyridinedicarboxylic acid (SGC - Diamond I04-1 fragment screening)
To be published
|
|
6P7B
 
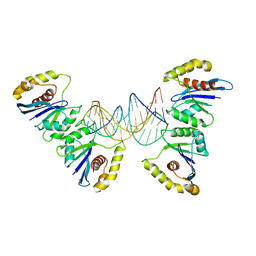 | | Crystal structure of Fowlpox virus resolvase and substrate Holliday junction DNA complex | | Descriptor: | DNA (29-MER), Holliday junction resolvase | | Authors: | Li, N, Shi, K, Rao, T, Banerjee, S, Aihara, H. | | Deposit date: | 2019-06-05 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.317 Å) | | Cite: | Structural insights into the promiscuous DNA binding and broad substrate selectivity of fowlpox virus resolvase.
Sci Rep, 10, 2020
|
|
6CS2
 
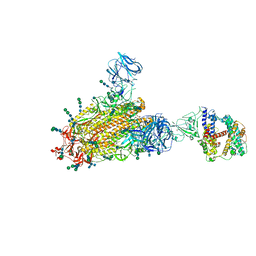 | | SARS Spike Glycoprotein - human ACE2 complex, Stabilized variant, all ACE2-bound particles | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
1A1Q
 
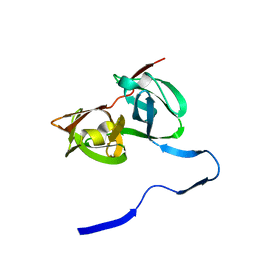 | | HEPATITIS C VIRUS NS3 PROTEINASE | | Descriptor: | NS3 PROTEINASE, ZINC ION | | Authors: | Love, R.A, Parge, H.E, Wickersham, J.A, Hostomsky, Z, Habuka, N, Moomaw, E.W, Adachi, T, Hostomska, Z. | | Deposit date: | 1997-12-12 | | Release date: | 1998-03-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of hepatitis C virus NS3 proteinase reveals a trypsin-like fold and a structural zinc binding site.
Cell(Cambridge,Mass.), 87, 1996
|
|
6FIU
 
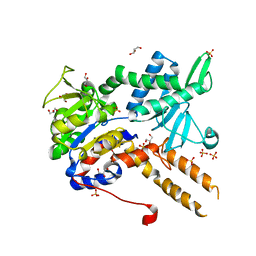 | |
1Q6V
 
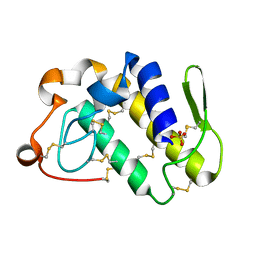 | | First crystal structure of a C49 monomer PLA2 from the venom of Daboia russelli pulchella at 1.8 A resolution | | Descriptor: | Phospholipase A2 VRV-PL-VIIIa, SULFATE ION | | Authors: | Singh, N, Pal, A, Jabeen, T, Sharma, S, Singh, T.P. | | Deposit date: | 2003-08-14 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | First crystal structure of a C49 PLA2 from the venom of Daboia russelli pulchella at 1.8A resolution
To be Published
|
|
6UI6
 
 | | HBV T=3 149C3A | | Descriptor: | Core protein | | Authors: | Wu, W, Watts, N.R, Cheng, N, Huang, R, Steven, A, Wingfield, P.T. | | Deposit date: | 2019-09-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Expression of quasi-equivalence and capsid dimorphism in the Hepadnaviridae.
Plos Comput.Biol., 16, 2020
|
|
5CUC
 
 | | Crystal structure of the bromodomain of bromodomain adjacent to zinc finger domain protein 2B (BAZ2B) in complex with N-Acetyl-2-phenylethylamine (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B, N-(2-phenylethyl)acetamide | | Authors: | Bradley, A, Pearce, N, Krojer, T, Ng, J, Talon, R, Vollmar, M, Jose, B, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the second bromodomain of bromodomain adjancent to zinc finger domain protein 2B (BAZ2B) in complex with N-Acetyl-2-phenylethylamine (SGC - Diamond I04-1 fragment screening)
To be published
|
|
