1FBV
 
 | | STRUCTURE OF A CBL-UBCH7 COMPLEX: RING DOMAIN FUNCTION IN UBIQUITIN-PROTEIN LIGASES | | Descriptor: | SIGNAL TRANSDUCTION PROTEIN CBL, SULFATE ION, UBIQUITIN-CONJUGATING ENZYME E12-18 KDA UBCH7, ... | | Authors: | Zheng, N, Wang, P, Jeffrey, P.D, Pavletich, N.P. | | Deposit date: | 2000-07-17 | | Release date: | 2000-08-30 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases.
Cell(Cambridge,Mass.), 102, 2000
|
|
1FI5
 
 | | NMR STRUCTURE OF THE C TERMINAL DOMAIN OF CARDIAC TROPONIN C BOUND TO THE N TERMINAL DOMAIN OF CARDIAC TROPONIN I. | | Descriptor: | CALCIUM ION, PROTEIN (TROPONIN C) | | Authors: | Gasmi-Seabrook, G.M, Howarth, J.W, Finley, N, Abusamhadneh, E, Gaponenko, V, Brito, R.M, Solaro, R.J, Rosevear, P.R. | | Deposit date: | 2000-08-03 | | Release date: | 2000-08-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the C-terminal domain of cardiac troponin C free and bound to the N-terminal domain of cardiac troponin I.
Biochemistry, 38, 1999
|
|
1FJJ
 
 | | CRYSTAL STRUCTURE OF E.COLI YBHB PROTEIN, A NEW MEMBER OF THE MAMMALIAN PEBP FAMILY | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HYPOTHETICAL 17.1 KDA PROTEIN IN MODC-BIOA INTERGENIC REGION | | Authors: | Serre, L, Pereira de Jesus, K, Benedetti, H, Bureaud, N, Schoentgen, F, Zelwer, C. | | Deposit date: | 2000-08-08 | | Release date: | 2001-07-18 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structures of YBHB and YBCL from Escherichia coli, two bacterial homologues to a Raf kinase inhibitor protein.
J.Mol.Biol., 310, 2001
|
|
1FJ5
 
 | | TAMOXIFEN-DNA ADDUCT | | Descriptor: | (Z)-2-[4-(1,2)-DIPHENYL-1-BUTENYL)-PHENOXY]-N,N-DIMETHYLETHANAMINIUM, DNA (5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3') | | Authors: | Shimotakahara, S, Gorin, A, Kolbanovskiy, A, Kettani, A, Hingerty, B.E, Amin, S, Broyde, S, Geacintov, N, Patel, D.J. | | Deposit date: | 2000-08-07 | | Release date: | 2000-09-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Accomodation of S-cis-tamoxifen-N(2)-guanine adduct within a bent and widened DNA minor groove.
J.Mol.Biol., 302, 2000
|
|
1CEE
 
 | | SOLUTION STRUCTURE OF CDC42 IN COMPLEX WITH THE GTPASE BINDING DOMAIN OF WASP | | Descriptor: | GTP-BINDING RHO-LIKE PROTEIN, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Abdul-Manan, N, Aghazadeh, B, Liu, G.A, Majumdar, A, Ouerfelli, O, Rosen, M.K. | | Deposit date: | 1999-03-08 | | Release date: | 1999-06-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of Cdc42 in complex with the GTPase-binding domain of the 'Wiskott-Aldrich syndrome' protein.
Nature, 399, 1999
|
|
1CZ6
 
 | | SOLUTION STRUCTURE OF ANDROCTONIN | | Descriptor: | PROTEIN (ANDROCTONIN) | | Authors: | Mandard, N, Vovelle, F. | | Deposit date: | 1999-09-01 | | Release date: | 2000-01-12 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Androctonin, a novel antimicrobial peptide from scorpion Androctonus australis: solution structure and molecular dynamics simulations in the presence of a lipid monolayer.
J.Biomol.Struct.Dyn., 17, 1999
|
|
1D28
 
 | |
1D79
 
 | | HIGH RESOLUTION REFINEMENT OF THE HEXAGONAL A-DNA OCTAMER D(GTGTACAC) AT 1.4 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*GP*TP*GP*TP*AP*CP*AP*C)-3') | | Authors: | Thota, N, Li, X.H, Bingman, C.A, Sundaralingam, M. | | Deposit date: | 1992-06-12 | | Release date: | 1993-04-15 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-resolution refinement of the hexagonal A-DNA octamer d(GTGTACAC) at 1.4 A.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1COA
 
 | | THE EFFECT OF CAVITY CREATING MUTATIONS IN THE HYDROPHOBIC CORE OF CHYMOTRYPSIN INHIBITOR 2 | | Descriptor: | CHYMOTRYPSIN INHIBITOR 2 | | Authors: | Jackson, S.E, Moracci, M, Elmasry, N, Johnson, C.M, Fersht, A.R. | | Deposit date: | 1993-05-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Effect of cavity-creating mutations in the hydrophobic core of chymotrypsin inhibitor 2.
Biochemistry, 32, 1993
|
|
1EAV
 
 | |
1EWV
 
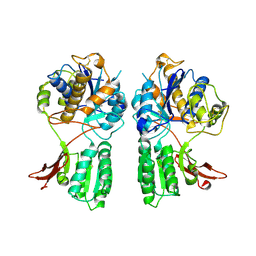 | | CRYSTAL STRUCTURE OF METABOTROPIC GLUTAMATE RECEPTOR SUBTYPE 1 LIGAND FREE FORM II | | Descriptor: | METABOTROPIC GLUTAMATE RECEPTOR SUBTYPE 1 | | Authors: | Kunishima, N, Shimada, Y, Tsuji, Y, Jingami, H, Morikawa, K. | | Deposit date: | 2000-04-27 | | Release date: | 2000-12-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor.
Nature, 407, 2000
|
|
1EWK
 
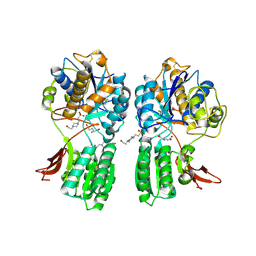 | | CRYSTAL STRUCTURE OF METABOTROPIC GLUTAMATE RECEPTOR SUBTYPE 1 COMPLEXED WITH GLUTAMATE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLUTAMIC ACID, ... | | Authors: | Kunishima, N, Shimada, Y, Jingami, H, Morikawa, K. | | Deposit date: | 2000-04-26 | | Release date: | 2000-12-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor.
Nature, 407, 2000
|
|
2D6Y
 
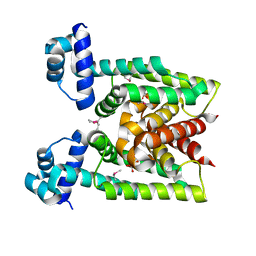 | | Crystal Structure of transcriptional factor SCO4008 from Streptomyces coelicolor A3(2) | | Descriptor: | L(+)-TARTARIC ACID, putative tetR family regulatory protein | | Authors: | Hayashi, T, Tanaka, Y, Sakai, N, Yao, M, Tamura, T, Tanaka, I. | | Deposit date: | 2005-11-15 | | Release date: | 2006-10-31 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | SCO4008, a Putative TetR Transcriptional Repressor from Streptomyces coelicolor A3(2), Regulates Transcription of sco4007 by Multidrug Recognition.
J.Mol.Biol., 425, 2013
|
|
1FI4
 
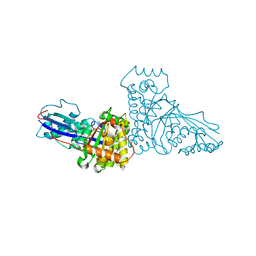 | | THE X-RAY CRYSTAL STRUCTURE OF MEVALONATE 5-DIPHOSPHATE DECARBOXYLASE AT 2.3 ANGSTROM RESOLUTION. | | Descriptor: | MEVALONATE 5-DIPHOSPHATE DECARBOXYLASE | | Authors: | Bonanno, J.B, Edo, C, Eswar, N, Pieper, U, Romanowski, M.J, Ilyin, V, Gerchman, S.E, Kycia, H, Studier, F.W, Sali, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2000-08-03 | | Release date: | 2001-03-21 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural genomics of enzymes involved in sterol/isoprenoid biosynthesis.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1FE1
 
 | | CRYSTAL STRUCTURE PHOTOSYSTEM II | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-2-METHYL-SUCCINIC ACID, CADMIUM ION, CHLOROPHYLL A, ... | | Authors: | Zouni, A, Witt, H.-T, Kern, J, Fromme, P, Krauss, N, Saenger, W, Orth, P. | | Deposit date: | 2000-07-20 | | Release date: | 2001-02-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of photosystem II from Synechococcus elongatus at 3.8 A resolution.
Nature, 409, 2001
|
|
1F9B
 
 | | MELANIN PROTEIN INTERACTION: X-RAY STRUCTURE OF THE COMPLEX OF MARE LACTOFERRIN WITH MELANIN MONOMERS | | Descriptor: | 3H-INDOLE-5,6-DIOL, BICARBONATE ION, FE (III) ION, ... | | Authors: | Kumar, S, Singh, T.P, Sharma, A.K, Singh, N, Raman, G. | | Deposit date: | 2000-07-10 | | Release date: | 2001-02-10 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Lactoferrin-melanin interaction and its possible implications in melanin polymerization: crystal structure of the complex formed between mare lactoferrin and melanin monomers at 2.7-A resolution.
Proteins, 45, 2001
|
|
2DM9
 
 | |
2DCH
 
 | | Crystal structure of archaeal intron-encoded homing endonuclease I-Tsp061I | | Descriptor: | CHLORIDE ION, SULFATE ION, putative homing endonuclease | | Authors: | Nakayama, H, Tsuge, H, Shimamura, T, Miyano, M, Nomura, N, Sako, Y. | | Deposit date: | 2006-01-06 | | Release date: | 2006-07-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure of a hyperthermophilic archaeal homing endonuclease, I-Tsp061I: contribution of cross-domain polar networks to thermostability.
J.Mol.Biol., 365, 2007
|
|
2DH1
 
 | | Crystal structure of peanut lectin lactose-azobenzene-4,4'-dicarboxylic acid-lactose complex | | Descriptor: | Galactose-binding lectin | | Authors: | Natchiar, S.K, Srinivas, O, Nivedita, M, Sagarika, D, Jayaraman, N, Surolia, A, Vijayan, M. | | Deposit date: | 2006-03-17 | | Release date: | 2006-08-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (7.65 Å) | | Cite: | Multivalency in lectins - A crystallographic, modelling and light-scattering study involving peanut lectin and a bivalent ligand
Curr.Sci., 90, 2006
|
|
1FOT
 
 | |
6AW6
 
 | | 1.70A resolution structure of catechol O-methyltransferase (COMT) L136M (rhombohedral form) from Nannospalax galili | | Descriptor: | Catechol O-methyltransferase, SULFATE ION | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Deng, Y, Hanzlik, R.P, Shams, I, Moskovitz, J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the catechol-o-methyl transferase (COMT) enzyme of the subterranean mole rat (Spalax) and the effect of L136M substitution
To be published
|
|
6AXD
 
 | |
6AP1
 
 | | Vps4p-Vta1p complex with peptide binding to the central pore of Vps4p | | Descriptor: | ACE-ASP-GLU-ILE-VAL-ASN-LYS-VAL-LEU-NH2, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Han, H, Monroe, N, Shen, P, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2017-08-16 | | Release date: | 2017-12-06 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The AAA ATPase Vps4 binds ESCRT-III substrates through a repeating array of dipeptide-binding pockets.
Elife, 6, 2017
|
|
6IC1
 
 | | urate oxidase under 90 bar of krypton | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 8-AZAXANTHINE, ACETATE ION, ... | | Authors: | Prange, T, Colloc'h, N, Carpentier, P. | | Deposit date: | 2018-12-01 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Comparative study of the effects of high hydrostatic pressure per se and high argon pressure on urate oxidase ligand stabilization
Acta Cryst. D, 78, 2022
|
|
6AW5
 
 | | 1.90A resolution structure of catechol O-methyltransferase (COMT) L136M (hexagonal form) from Nannospalax galili | | Descriptor: | CHLORIDE ION, Catechol O-methyltransferase, GLYCEROL, ... | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Deng, Y, Hanzlik, R.P, Shams, I, Moskovitz, J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the catechol-o-methyl transferase (COMT) enzyme of the subterranean mole rat (Spalax) and the effect of L136M substitution
To be published
|
|
