1UHR
 
 | | Solution structure of the SWIB domain of mouse BRG1-associated factor 60a | | Descriptor: | SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily D member 1 | | Authors: | Yamada, K, Saito, K, Nameki, N, Inoue, M, Koshiba, S, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Matsuda, T, Seki, E, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-09 | | Release date: | 2004-08-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SWIB domain of mouse BRG1-associated factor 60a
To be Published
|
|
6WS1
 
 | | Crystal structure of human phenylethanolamine N-methyltransferase (PNMT) in complex with (2S)-2-amino-4-((((2R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl)(3-(7,8-dichloro-1,2,3,4-tetrahydroisoquinolin-4-yl)propyl)amino)butanoic acid and AdoHcy (SAH) | | Descriptor: | 1,2-ETHANEDIOL, 5'-([(3S)-3-amino-3-carboxypropyl]{3-[(4R)-7,8-dichloro-1,2,3,4-tetrahydroisoquinolin-4-yl]propyl}amino)-5'-deoxyadenosine, CADMIUM ION, ... | | Authors: | Harijan, R.K, Mahmoodi, N, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2020-04-30 | | Release date: | 2020-08-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Transition-State Analogues of PhenylethanolamineN-Methyltransferase.
J.Am.Chem.Soc., 142, 2020
|
|
6WOS
 
 | |
8GNN
 
 | | Crystal structure of the human RAD9-RAD1-HUS1-RAD17 complex | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1, Cell cycle checkpoint protein RAD17, ... | | Authors: | Hara, K, Nagata, K, Iida, N, Hashimoto, H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.119 Å) | | Cite: | The 9-1-1 DNA clamp subunit RAD1 forms specific interactions with clamp loader RAD17, revealing functional implications for binding-protein RHINO.
J.Biol.Chem., 299, 2023
|
|
6WV7
 
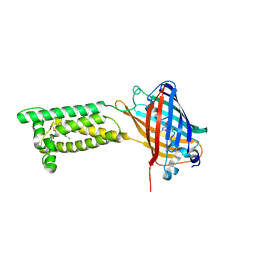 | | Human VKOR with Chlorophacinone | | Descriptor: | Chlorophacinone, Vitamin K epoxide reductase, termini restrained by green fluorescent protein | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.483 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
6WVT
 
 | | Structural basis of alphaE-catenin - F-actin catch bond behavior | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Xu, X.P, Pokutta, S, Torres, M, Swift, M.F, Hanein, D, Volkmann, N, Weis, W.I. | | Deposit date: | 2020-05-06 | | Release date: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural basis of alpha E-catenin-F-actin catch bond behavior.
Elife, 9, 2020
|
|
6WF1
 
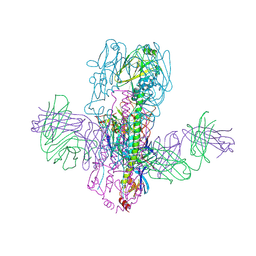 | | Crystal Structure of Broadly Neutralizing Antibody 3I14 Bound to the Influenza A H10 Hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, Hemagglutinin HA2 chain, ... | | Authors: | Harshbarger, W.D, Lockbaum, G.J, Deming, D.T, Attatippaholkun, N, Schiffer, C.A, Marasco, W.A. | | Deposit date: | 2020-04-03 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.19 Å) | | Cite: | Unique structural solution from a V H 3-30 antibody targeting the hemagglutinin stem of influenza A viruses.
Nat Commun, 12, 2021
|
|
6WNV
 
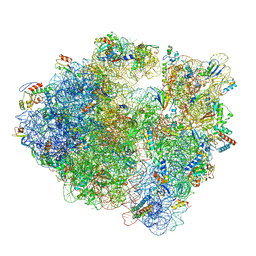 | | 70S ribosome without free 5S rRNA and with a perturbed PTC | | Descriptor: | 16S ribosomal RNA, 23s-5s joint ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Korostelev, A.A, Mankin, A.S, Huang, S, Aleksashin, N.A, Klepacki, D, Reier, K, Kefi, A, Szal, A, Remme, J, Jaeger, L, Vazquez-Laslop, N. | | Deposit date: | 2020-04-23 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Ribosome engineering reveals the importance of 5S rRNA autonomy for ribosome assembly.
Nat Commun, 11, 2020
|
|
1UZV
 
 | | High affinity fucose binding of Pseudomonas aeruginosa lectin II: 1.0 A crystal structure of the complex | | Descriptor: | CALCIUM ION, PSEUDOMONAS AERUGINOSA LECTIN II, SULFATE ION, ... | | Authors: | Mitchell, E, Sabin, C.D, Snajdrova, L, Budova, M, Perret, S, Gautier, C, Gilboa-Garber, N, Koca, J, Wimmerova, M, Imberty, A. | | Deposit date: | 2004-03-17 | | Release date: | 2004-12-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High Affinity Fucose Binding of Pseudomonas Aeruginosa Lectin Pa-Iil: 1.0 A Resolution Crystal Structure of the Complex Combined with Thermodynamics and Computational Chemistry Approaches.
Proteins, 58, 2005
|
|
6WVV
 
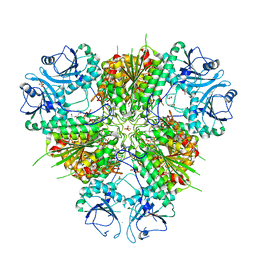 | | Plasmodium vivax M17 leucyl aminopeptidase | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, M17 leucyl aminopeptidase, ... | | Authors: | Malcolm, T.R, Drinkwater, N, McGowan, S. | | Deposit date: | 2020-05-07 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Active site metals mediate an oligomeric equilibrium in Plasmodium M17 aminopeptidases.
J.Biol.Chem., 296, 2020
|
|
1UJV
 
 | |
1UJS
 
 | |
1UJY
 
 | | Solution structure of SH3 domain in Rac/Cdc42 guanine nucleotide exchange factor(GEF) 6 | | Descriptor: | Rho guanine nucleotide exchange factor 6 | | Authors: | He, F, Muto, Y, Uda, H, Koshiba, S, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Ohara, O, Nagase, T, Kikuno, R, Nagayama, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-08-12 | | Release date: | 2004-02-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of SH3 domain in Rac/Cdc42 guanine nucleotide exchange factor(GEF) 6
To be Published
|
|
6WH8
 
 | | The structure of NTMT1 in complex with compound BM-30 | | Descriptor: | 4HP-PRO-LYS-ARG-NH2, BM-30, N-terminal Xaa-Pro-Lys N-methyltransferase 1, ... | | Authors: | Noinaj, N, Chen, D, Huang, R. | | Deposit date: | 2020-04-07 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.729 Å) | | Cite: | Selective Peptidomimetic Inhibitors of NTMT1/2: Rational Design, Synthesis, Characterization, and Crystallographic Studies.
J.Med.Chem., 63, 2020
|
|
1UJU
 
 | |
1UJT
 
 | |
6WHY
 
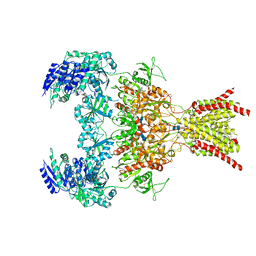 | | GluN1b-GluN2B NMDA receptor in complex with GluN1 antagonist L689,560, class 1 | | Descriptor: | (2R,4S)-5,7-dichloro-4-[(phenylcarbamoyl)amino]-1,2,3,4-tetrahydroquinoline-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chou, T, Tajima, N, Furukawa, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-07-15 | | Last modified: | 2020-08-05 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
1BWV
 
 | | Activated Ribulose 1,5-Bisphosphate Carboxylase/Oxygenase (RUBISCO) Complexed with the Reaction Intermediate Analogue 2-Carboxyarabinitol 1,5-Bisphosphate | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, PROTEIN (RIBULOSE BISPHOSPHATE CARBOXYLASE) | | Authors: | Sugawara, H, Yamamoto, H, Shibata, N, Inoue, T, Miyake, C, Yokota, A, Kai, Y. | | Deposit date: | 1998-09-29 | | Release date: | 1999-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of carboxylase reaction-oriented ribulose 1, 5-bisphosphate carboxylase/oxygenase from a thermophilic red alga, Galdieria partita.
J.Biol.Chem., 274, 1999
|
|
1R56
 
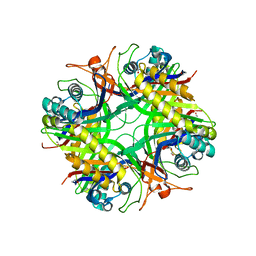 | | UNCOMPLEXED URATE OXIDASE FROM ASPERGILLUS FLAVUS | | Descriptor: | DI(HYDROXYETHYL)ETHER, Uricase | | Authors: | Retailleau, P, Colloc'h, N, Prange, T. | | Deposit date: | 2003-10-09 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Complexed and ligand-free high-resolution structures of urate oxidase (Uox) from Aspergillus flavus: a reassignment of the active-site binding mode.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1C4X
 
 | | 2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE (BPHD) FROM RHODOCOCCUS SP. STRAIN RHA1 | | Descriptor: | PROTEIN (2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE) | | Authors: | Nandhagopal, N, Senda, T, Mitsui, Y. | | Deposit date: | 1999-09-30 | | Release date: | 1999-10-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three-Dimensional Structure of Microbial 2-Hydroxyl-6-Oxo-6-Phenylhexa-2,4- Dienoic Acid (Hpda) Hydrolase (Bphd Enzyme) from Rhodococcus Sp. Strain Rha1, in the Pcb Degradation Pathway
Proc.Jpn.Acad.,Ser.B, 73, 1997
|
|
3W7Y
 
 | |
3W80
 
 | |
1C8Q
 
 | |
1R51
 
 | | URATE OXIDASE FROM ASPERGILLUS FLAVUS COMPLEXED WITH ITS INHIBITOR 8-AZAXANTHIN | | Descriptor: | 8-AZAXANTHINE, CYSTEINE, Uricase | | Authors: | Prange, T, Retailleau, P, Colloc'h, N. | | Deposit date: | 2003-10-09 | | Release date: | 2004-03-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Complexed and ligand-free high-resolution structures of urate oxidase (Uox) from Aspergillus flavus: a reassignment of the active-site binding mode.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1V5M
 
 | | Solution Structure of the Pleckstrin Homology Domain of Mouse APS | | Descriptor: | SH2 and PH domain-containing adapter protein APS | | Authors: | Li, H, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-25 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Pleckstrin Homology Domain of Mouse APS
To be Published
|
|
