6IMW
 
 | | The complex structure of endo-beta-1,2-glucanase mutant (E262Q) from Talaromyces funiculosus with beta-1,2-glucan | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, Endo-beta-1,2-glucanase, ... | | Authors: | Tanaka, N, Nakajima, M, Narukawa-Nara, M, Matsunaga, H, Kamisuki, S, Aramasa, H, Takahashi, Y, Sugimoto, N, Abe, K, Miyanaga, A, Yamashita, T, Sugawara, F, Kamakura, T, Komba, S, Nakai, H, Taguchi, H. | | Deposit date: | 2018-10-23 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification, characterization, and structural analyses of a fungal endo-beta-1,2-glucanase reveal a new glycoside hydrolase family.
J.Biol.Chem., 294, 2019
|
|
1X57
 
 | | Solution structures of the HTH domain of human EDF-1 protein | | Descriptor: | Endothelial differentiation-related factor 1 | | Authors: | Nameki, N, Sato, M, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-15 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the HTH domain of human EDF-1 protein
To be Published
|
|
6TWR
 
 | |
6J0B
 
 | | Cryo-EM Structure of an Extracellular Contractile Injection System, PVC sheath-tube complex in extended state | | Descriptor: | Pvc1, Pvc2 | | Authors: | Jiang, F, Li, N, Wang, X, Cheng, J, Huang, Y, Yang, Y, Yang, J, Cai, B, Wang, Y, Jin, Q, Gao, N. | | Deposit date: | 2018-12-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM Structure and Assembly of an Extracellular Contractile Injection System.
Cell, 177, 2019
|
|
6J0C
 
 | | Cryo-EM Structure of an Extracellular Contractile Injection System, PVC sheath complex in contracted state | | Descriptor: | Pvc2 | | Authors: | Jiang, F, Li, N, Wang, X, Cheng, J, Huang, Y, Yang, Y, Yang, J, Cai, B, Wang, Y, Jin, Q, Gao, N. | | Deposit date: | 2018-12-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM Structure and Assembly of an Extracellular Contractile Injection System.
Cell, 177, 2019
|
|
6RNH
 
 | | Structure of C-terminal truncated Plasmodium falciparum IMP-nucleotidase | | Descriptor: | GLYCEROL, IMP-specific 5'-nucleotidase, putative | | Authors: | Carrique, L, Ballut, L, Violot, S, Aghajari, N. | | Deposit date: | 2019-05-08 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure and catalytic regulation of Plasmodium falciparum IMP specific nucleotidase.
Nat Commun, 11, 2020
|
|
1WH8
 
 | | Solution structure of the third CUT domain of human Homeobox protein Cux-2 | | Descriptor: | Homeobox protein Cux-2 | | Authors: | Nameki, N, Tochio, N, Koshiba, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the third CUT domain of human Homeobox protein Cux-2
To be Published
|
|
6RN1
 
 | | Structure of N-terminal truncated Plasmodium falciparum IMP-nucleotidase | | Descriptor: | IMP-specific 5'-nucleotidase, putative | | Authors: | Carrique, L, Ballut, L, Violot, S, Aghajari, N. | | Deposit date: | 2019-05-07 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and catalytic regulation of Plasmodium falciparum IMP specific nucleotidase.
Nat Commun, 11, 2020
|
|
6J4H
 
 | | Crystal Structure of maltotriose-complex of PulA-G680L mutant from Klebsiella pneumoniae | | Descriptor: | CALCIUM ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2019-01-09 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Relationship between the induced-fit loop and the activity of Klebsiella pneumoniae pullulanase.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6UXB
 
 | |
6J0F
 
 | | Cryo-EM Structure of an Extracellular Contractile Injection System, PVC sheath/tube terminator in extended state | | Descriptor: | Pvc1, Pvc16 | | Authors: | Jiang, F, Li, N, Wang, X, Cheng, J, Huang, Y, Yang, Y, Yang, J, Cai, B, Wang, Y, Jin, Q, Gao, N. | | Deposit date: | 2018-12-24 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM Structure and Assembly of an Extracellular Contractile Injection System.
Cell, 177, 2019
|
|
6J35
 
 | | Crystal structure of ligand-free of PulA-G680L mutant from Klebsiella pneumoniae | | Descriptor: | CALCIUM ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2019-01-09 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.839 Å) | | Cite: | Relationship between the induced-fit loop and the activity of Klebsiella pneumoniae pullulanase.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6RMO
 
 | | Structure of Plasmodium falciparum IMP-nucleotidase | | Descriptor: | IMP-specific 5'-nucleotidase, putative | | Authors: | Carrique, L, Ballut, L, Violot, S, Aghajari, N. | | Deposit date: | 2019-05-07 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and catalytic regulation of Plasmodium falciparum IMP specific nucleotidase.
Nat Commun, 11, 2020
|
|
8E37
 
 | | Structure of Campylobacter concisus wild-type SeMet PglC | | Descriptor: | N,N'-diacetylbacilliosaminyl-1-phosphate transferase | | Authors: | Vuksanovic, N, Ray, L.C, Imperiali, B, Allen, K.N. | | Deposit date: | 2022-08-16 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Synergistic computational and experimental studies of a phosphoglycosyl transferase membrane/ligand ensemble.
J.Biol.Chem., 299, 2023
|
|
4BQJ
 
 | | structure of HSP90 with an inhibitor bound | | Descriptor: | 5-[2,4-dihydroxy-6-(4-nitrophenoxy)phenyl]-N-ethyl-1,2-oxazole-3-carboxamide, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Casale, E, Brasca, M.G, Mantegani, S, Amboldi, N, Bindi, S, Caronni, D, Ceccarelli, W, Colombo, N, DePonti, A, Donati, D, Ermoli, A, Fachin, G, Felder, E.R, Ferguson, R.D, Fiorelli, C, Guanci, M, Isacchi, A, Pesenti, E, Polucci, P, Riceputi, L, Sola, F, Visco, C, Zuccotto, F, Fogliatto, G. | | Deposit date: | 2013-05-30 | | Release date: | 2013-10-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Nms-E973 as Novel, Selective and Potent Inhibitor of Heat Shock Protein 90 (Hsp90).
Bioorg.Med.Chem., 21, 2013
|
|
6RME
 
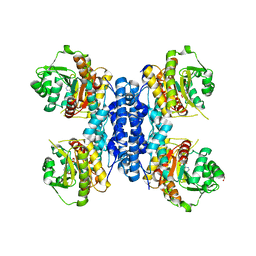 | | Structure of IMP bound Plasmodium falciparum IMP-nucleotidase mutant D172N | | Descriptor: | GLYCEROL, IMP-specific 5'-nucleotidase, putative, ... | | Authors: | Carrique, L, Ballut, L, Violot, S, Aghajari, N. | | Deposit date: | 2019-05-06 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and catalytic regulation of Plasmodium falciparum IMP specific nucleotidase.
Nat Commun, 11, 2020
|
|
6XQK
 
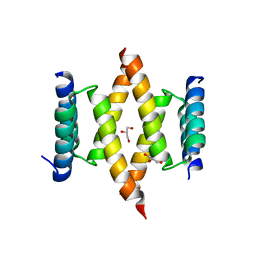 | | Crystal structure of the D/D domain of PKA from S. cerevisiae | | Descriptor: | CHLORIDE ION, GLYCEROL, cAMP-dependent protein kinase regulatory subunit | | Authors: | Larrieux, N, Gonzalez Bardeci, N, Trajtenberg, F, Buschiazzo, A. | | Deposit date: | 2020-07-09 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The crystal structure of yeast regulatory subunit reveals key evolutionary insights into Protein Kinase A oligomerization.
J.Struct.Biol., 213, 2021
|
|
1WH6
 
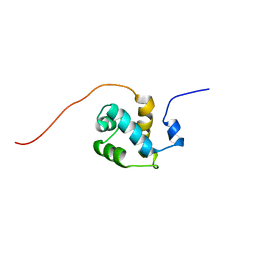 | | Solution structure of the second CUT domain of human Homeobox protein Cux-2 | | Descriptor: | Homeobox protein Cux-2 | | Authors: | Nameki, N, Tochio, N, Koshiba, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second CUT domain of human Homeobox protein Cux-2
To be Published
|
|
6J0N
 
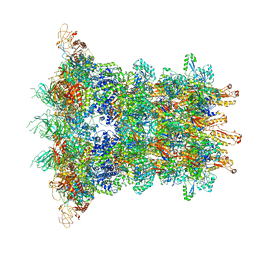 | | Cryo-EM Structure of an Extracellular Contractile Injection System, baseplate in extended state, refined in C6 symmetry | | Descriptor: | Pvc1, Pvc11, Pvc12, ... | | Authors: | Jiang, F, Li, N, Wang, X, Cheng, J, Huang, Y, Yang, Y, Yang, J, Cai, B, Wang, Y, Jin, Q, Gao, N. | | Deposit date: | 2018-12-25 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM Structure and Assembly of an Extracellular Contractile Injection System.
Cell, 177, 2019
|
|
6J0M
 
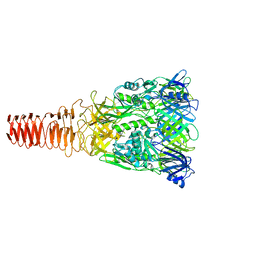 | | Cryo-EM Structure of an Extracellular Contractile Injection System, PVC baseplate in extended state (reconstructed with C3 symmetry) | | Descriptor: | Pvc8 | | Authors: | Jiang, F, Li, N, Wang, X, Cheng, J, Huang, Y, Yang, Y, Yang, J, Cai, B, Wang, Y, Jin, Q, Gao, N. | | Deposit date: | 2018-12-24 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM Structure and Assembly of an Extracellular Contractile Injection System.
Cell, 177, 2019
|
|
1AJ1
 
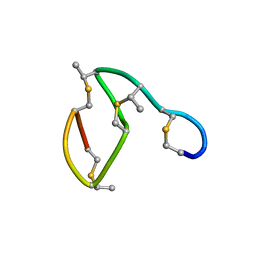 | | NMR STRUCTURE OF THE LANTIBIOTIC ACTAGARDINE | | Descriptor: | LANTIBIOTIC ACTAGARDINE | | Authors: | Zimmermann, N, Jung, G. | | Deposit date: | 1997-05-14 | | Release date: | 1997-10-15 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional solution structure of the lantibiotic murein-biosynthesis-inhibitor actagardine determined by NMR.
Eur.J.Biochem., 246, 1997
|
|
6UXA
 
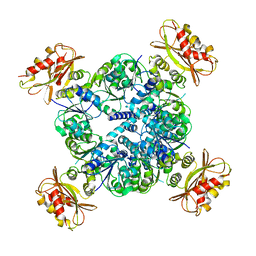 | |
4C7O
 
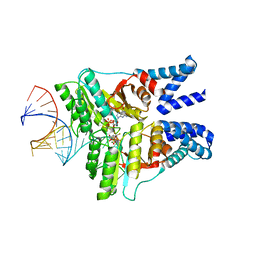 | | The structural basis of FtsY recruitment and GTPase activation by SRP RNA | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SIGNAL RECOGNITION PARTICLE PROTEIN, ... | | Authors: | Voigts-Hoffmann, F, Schmitz, N, Shen, K, Shan, S.O, Ataide, S.F, Ban, N. | | Deposit date: | 2013-09-23 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structural Basis of Ftsy Recruitment and Gtpase Activation by Srp RNA
Mol.Cell, 52, 2013
|
|
6J33
 
 | | Crystal structure of ligand-free of PulA from Klebsiella pneumoniae | | Descriptor: | ACETATE ION, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2019-01-09 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.297 Å) | | Cite: | Relationship between the induced-fit loop and the activity of Klebsiella pneumoniae pullulanase.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6JSG
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(4S)-2-amino-4-methyl-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-5-chloropyridine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Fujimoto, K, Matsuoka, E, Asada, N, Tadano, G, Yamamoto, T, Nakahara, K, Fuchino, K, Ito, H, Kanegawa, N, Moechars, D, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Selective beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors: Targeting the Flap to Gain Selectivity over BACE2.
J.Med.Chem., 62, 2019
|
|
