6PW7
 
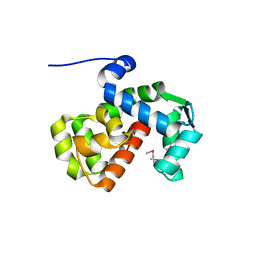 | | X-ray crystal structure of C. elegans STIM EF-SAM domain | | Descriptor: | CALCIUM ION, Stromal interaction molecule 1 | | Authors: | Enomoto, M, Nishikawa, T, Back, S.I, Ishiyama, N, Zheng, L, Stathopulos, P.B, Ikura, M. | | Deposit date: | 2019-07-22 | | Release date: | 2019-11-13 | | Last modified: | 2020-02-12 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Coordination of a Single Calcium Ion in the EF-hand Maintains the Off State of the Stromal Interaction Molecule Luminal Domain.
J.Mol.Biol., 432, 2020
|
|
6PT2
 
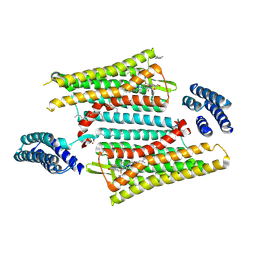 | | Crystal structure of the active delta opioid receptor in complex with the peptide agonist KGCHM07 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, Delta opioid receptor, ... | | Authors: | Claff, T, Yu, J, Blais, V, Patel, N, Martin, C, Wu, L, Han, G.W, Holleran, B.J, Van der Poorten, O, Hanson, M.A, Sarret, P, Gendron, L, Cherezov, V, Katritch, V, Ballet, S, Liu, Z, Muller, C.E, Stevens, R.C. | | Deposit date: | 2019-07-14 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Elucidating the active delta-opioid receptor crystal structure with peptide and small-molecule agonists.
Sci Adv, 5, 2019
|
|
6PU9
 
 | | Crystal Structure of the Type B Chloramphenicol O-Acetyltransferase from Vibrio vulnificus | | Descriptor: | 1,2-ETHANEDIOL, Acetyltransferase, CHLORIDE ION | | Authors: | Kim, Y, Maltseva, N, Mulligan, R, Grimshaw, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-17 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional characterization of three Type B and C chloramphenicol acetyltransferases from Vibrio species.
Protein Sci., 29, 2020
|
|
6PUA
 
 | | The 2.0 A Crystal Structure of the Type B Chloramphenicol Acetyltransferase from Vibrio cholerae | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Kim, Y, Maltseva, N, Stam, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional characterization of three Type B and C chloramphenicol acetyltransferases from Vibrio species.
Protein Sci., 29, 2020
|
|
5NL9
 
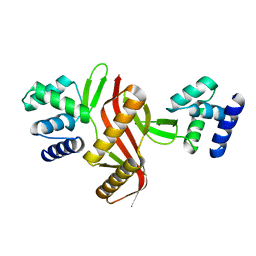 | | Crystal structure of a peroxide stress regulator from Leptospira interrogans | | Descriptor: | POTASSIUM ION, Transcriptional regulator (FUR family), UNKNOWN ATOM OR ION, ... | | Authors: | Saul, F.A, Haouz, A, Picardeau, M, Benaroudj, N. | | Deposit date: | 2017-04-04 | | Release date: | 2017-11-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of the Leptospira interrogans peroxide stress regulator (PerR), an atypical PerR devoid of a structural metal-binding site.
J. Biol. Chem., 293, 2018
|
|
6PVB
 
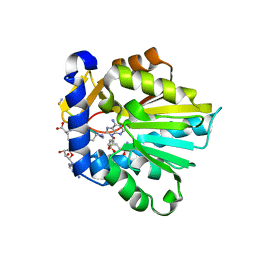 | | The structure of NTMT1 in complex with compound 6 | | Descriptor: | AMINO GROUP-()-(2~{S})-2-azanylpropanal-()-ISOLEUCINE-()-ARGININE-()-LYSINE-()-PROLINE-()-AMINO-ACETALDEHYDE-()-9-(5-{[(3S)-3-amino-3-carboxypropyl](pentyl)amino}-5-deoxy-beta-L-arabinofuranosyl)-9H-purin-6-amine, N-terminal Xaa-Pro-Lys N-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Noinaj, N, Chen, D, Huang, R. | | Deposit date: | 2019-07-20 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Probing the Plasticity in the Active Site of Protein N-terminal Methyltransferase 1 Using Bisubstrate Analogues.
J.Med.Chem., 63, 2020
|
|
5NCO
 
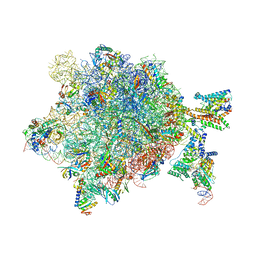 | | Quaternary complex between SRP, SR, and SecYEG bound to the translating ribosome | | Descriptor: | 23S rRNA, 4.5S SRP RNA (Ffs), 50S ribosomal protein L10, ... | | Authors: | Jomaa, A, Hwang Fu, Y, Boerhinger, D, Leibundgut, M, Shan, S.O, Ban, N. | | Deposit date: | 2017-03-06 | | Release date: | 2017-05-24 | | Last modified: | 2018-03-28 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of the quaternary complex between SRP, SR, and translocon bound to the translating ribosome.
Nat Commun, 8, 2017
|
|
5N9S
 
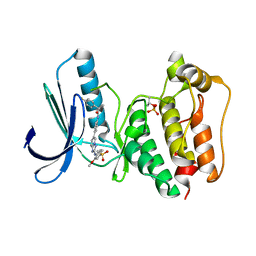 | | TTK kinase domain in complex with BAY 1161909 | | Descriptor: | (2~{R})-2-(4-fluorophenyl)-~{N}-[4-[2-[(2-methoxy-4-methylsulfonyl-phenyl)amino]-[1,2,4]triazolo[1,5-a]pyridin-6-yl]phenyl]propanamide, Dual specificity protein kinase TTK | | Authors: | Uitdehaag, J, Willemsen-Seegers, N, de Man, J, Buijsman, R.C, Zaman, G.J.R. | | Deposit date: | 2017-02-27 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Target Residence Time-Guided Optimization on TTK Kinase Results in Inhibitors with Potent Anti-Proliferative Activity.
J. Mol. Biol., 429, 2017
|
|
5NB7
 
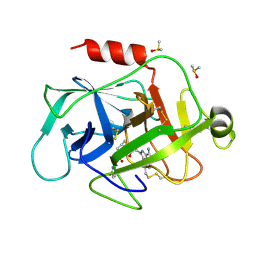 | | Complement factor D | | Descriptor: | 1-[2-[(1~{R},3~{S},5~{R})-3-[(6-bromanylpyridin-2-yl)carbamoyl]-2-azabicyclo[3.1.0]hexan-2-yl]-2-oxidanylidene-ethyl]indazole-3-carboxamide, Complement factor D, DIMETHYL SULFOXIDE | | Authors: | Mac Sweeney, A, Ostermann, N. | | Deposit date: | 2017-03-01 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Discovery of Highly Potent and Selective Small-Molecule Reversible Factor D Inhibitors Demonstrating Alternative Complement Pathway Inhibition in Vivo.
J. Med. Chem., 60, 2017
|
|
5ND1
 
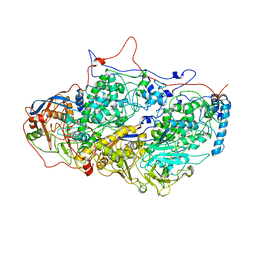 | | Viral evolution results in multiple, surface-allocated enzymatic activities in a fungal double-stranded RNA virus | | Descriptor: | Capsid protein | | Authors: | Mata, C.P, Luque, D, Gomez Blanco, J, Rodriguez, J.M, Suzuki, N, Ghabrial, S.A, Carrascosa, J.L, Trus, B.L, Caston, J.R. | | Deposit date: | 2017-03-07 | | Release date: | 2017-11-29 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Acquisition of functions on the outer capsid surface during evolution of double-stranded RNA fungal viruses.
PLoS Pathog., 13, 2017
|
|
6Q7U
 
 | |
5MZO
 
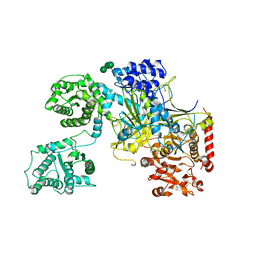 | | UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum (open conformation) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roversi, P, Caputo, A.T, Hill, J, Alonzi, D.S, Zitzmann, N. | | Deposit date: | 2017-02-01 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Interdomain conformational flexibility underpins the activity of UGGT, the eukaryotic glycoprotein secretion checkpoint.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5N2J
 
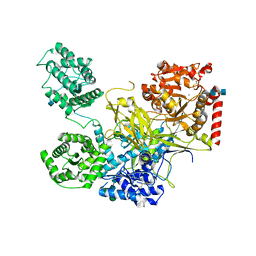 | | UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum (closed form) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roversi, P, Caputo, A.T, Hill, J, Alonzi, D.S, Zitzmann, N. | | Deposit date: | 2017-02-07 | | Release date: | 2017-07-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Interdomain conformational flexibility underpins the activity of UGGT, the eukaryotic glycoprotein secretion checkpoint.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6PXG
 
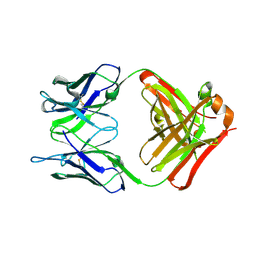 | |
5N2R
 
 | | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with PSB36 at 2.8A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-butyl-3-[(~{E})-3-oxidanylprop-1-enyl]-8-[(1~{R},5~{S})-3-tricyclo[3.3.1.0^{3,7}]nonanyl]-7~{H}-purine-2,6-dione, Adenosine receptor A2a,Soluble cytochrome b562,ADENOSINE RECEPTOR A2A SOLUBLE CYTOCHROME B562 ADENOSINE RECEPTOR A2A,Adenosine receptor A2a, ... | | Authors: | Cheng, R.K.Y, Segala, E, Robertson, N, Deflorian, F, Dore, A.S, Errey, J.C, Fiez-Vandal, C, Marshall, F.H, Cooke, R.M. | | Deposit date: | 2017-02-08 | | Release date: | 2017-07-26 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Human A1 and A2A Adenosine Receptors with Xanthines Reveal Determinants of Selectivity.
Structure, 25, 2017
|
|
6QE2
 
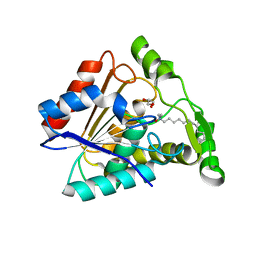 | | Crystal structure of Paleococcus ferrophilus monoacylglycerol lipase. | | Descriptor: | GLYCEROL, LAURYL DIMETHYLAMINE-N-OXIDE, Monoacylglycerol lipase | | Authors: | Labar, G, Demarez, M, Brandt, N, Wouters, J, Leherte, F. | | Deposit date: | 2019-01-04 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7461 Å) | | Cite: | Structure and Dynamics of an Archeal Monoglyceride Lipase from Palaeococcus ferrophilus as Revealed by Crystallography and In Silico Analysis.
Biomolecules, 11, 2021
|
|
6PT7
 
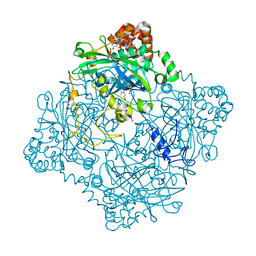 | | Structure of KatE1 catalase from Acinetobacter sp. Ver3 | | Descriptor: | CHLORIDE ION, Catalase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Gonzalez, J.M, Sartorio, M.G, Cortez, N. | | Deposit date: | 2019-07-15 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and functional properties of the cold-adapted catalase from Acinetobacter sp. Ver3 native to the Atacama plateau in northern Argentina
Acta Crystallogr.,Sect.D, D77, 2021
|
|
5NE0
 
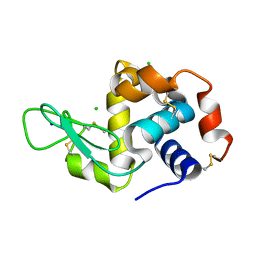 | | Room temperature in-situ structure of hen egg-white lysozyme from crystals enclosed between ultrathin silicon nitride membranes | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Martiel, I, Opara, N, Arnold, S.A, Braun, T, Stahlberg, H, Makita, M, David, C, Padeste, C. | | Deposit date: | 2017-03-09 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Direct protein crystallization on ultrathin membranes for diffraction measurements at X-ray free-electron lasers.
J.Appl.Crystallogr., 50, 2017
|
|
5MZJ
 
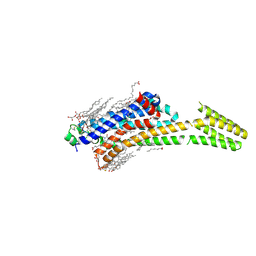 | | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with theophylline at 2.0A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Cheng, R.K.Y, Segala, E, Robertson, N, Deflorian, F, Dore, A.S, Errey, J.C, Fiez-Vandal, C, Marshall, F.H, Cooke, R.M. | | Deposit date: | 2017-01-31 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Human A1 and A2A Adenosine Receptors with Xanthines Reveal Determinants of Selectivity.
Structure, 25, 2017
|
|
5N2M
 
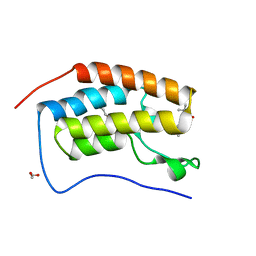 | | Crystal structure of the first bromodomain of human BRD4 in complex with a tetrahydroquinoline analogue | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, propan-2-yl ~{N}-[(2~{S},4~{R})-6-(3-acetamidophenyl)-1-ethanoyl-2-methyl-3,4-dihydro-2~{H}-quinolin-4-yl]carbamate | | Authors: | Tallant, C, Slavish, P.J, Siejka, P, Bharatham, N, Shadrick, W.R, Chai, S, Young, B.M, Boyd, V.A, Heroven, C, Wiggers, H.J, Picaud, S, Fedorov, O, Krojer, T, Chen, T, Lee, R.E, Guy, R.K, Shelat, A.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-02-07 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of the first bromodomain of human BRD4 in complex with a tetrahydroquinoline analogue
To Be Published
|
|
6QCY
 
 | | MloK1 model from single particle analysis of 2D crystals, class 2 (intermediate conformation) | | Descriptor: | Cyclic nucleotide-gated potassium channel mll3241, POTASSIUM ION | | Authors: | Righetto, R, Biyani, N, Kowal, J, Chami, M, Stahlberg, H. | | Deposit date: | 2018-12-31 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Retrieving high-resolution information from disordered 2D crystals by single-particle cryo-EM.
Nat Commun, 10, 2019
|
|
6Q81
 
 | | Structure of P-glycoprotein(ABCB1) in the post-hydrolytic state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, P-glycoprotein (ABCB1) | | Authors: | Ford, R.C, Thonghin, N, Collins, R.F, Barbieri, A, Shafi, T, Siebert, A. | | Deposit date: | 2018-12-13 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Novel features in the structure of P-glycoprotein (ABCB1) in the post-hydrolytic state as determined at 7.9 angstrom resolution.
Bmc Struct.Biol., 18, 2018
|
|
5N6U
 
 | |
7F1D
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(4R,5R,6R)-2-amino-5-fluoro-4,6-dimethyl-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-2H,3H-[1,4]dioxino[2,3-c]pyridine-7-carboxamide | | Descriptor: | Beta-secretase 1, IODIDE ION, N-[3-[(4R,5R,6R)-2-azanyl-5-fluoranyl-4,6-dimethyl-5,6-dihydro-1,3-thiazin-4-yl]-4-fluoranyl-phenyl]-2,3-dihydro-[1,4]dioxino[2,3-c]pyridine-7-carboxamide | | Authors: | Ueno, T, Matsuoka, E, Asada, N, Yamamoto, S, Kanegawa, N, Ito, M, Ito, H, Moechars, D, Rombouts, F.J.R, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2021-06-09 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Extremely Selective Fused Pyridine-Derived beta-Site Amyloid Precursor Protein-Cleaving Enzyme (BACE1) Inhibitors with High In Vivo Efficacy through 10s Loop Interactions.
J.Med.Chem., 64, 2021
|
|
5NA0
 
 | | TTK kinase domain in complex with a PEG-linked pyrimido-indolizine | | Descriptor: | Dual specificity protein kinase TTK, ~{N}-[3,5-diethyl-1-[2-[2-(2-methoxyethoxy)ethoxy]ethyl]pyrazol-4-yl]-2-[[2-methoxy-4-[4-(2-methoxyethanoyl)piperazin-1-yl]phenyl]amino]-5,6-dihydropyrimido[4,5-e]indolizine-7-carboxamide | | Authors: | Uitdehaag, J, Willemsen-Seegers, N, de Man, J, Buijsman, R.C, Zaman, G.J.R. | | Deposit date: | 2017-02-27 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Target Residence Time-Guided Optimization on TTK Kinase Results in Inhibitors with Potent Anti-Proliferative Activity.
J. Mol. Biol., 429, 2017
|
|
