9FVE
 
 | |
6G32
 
 | | Crystal structure of human geranylgeranyl diphosphate synthase mutant D188Y | | Descriptor: | GLYCEROL, Geranylgeranyl pyrophosphate synthase | | Authors: | Lisnyansky, M, Kapelushnik, N, Ben-Bassat, A, Marom, M, Loewenstein, A, Khananshvili, D, Giladi, M, Haitin, Y. | | Deposit date: | 2018-03-24 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.281 Å) | | Cite: | Reduced Activity of Geranylgeranyl Diphosphate Synthase Mutant Is Involved in Bisphosphonate-Induced Atypical Fractures.
Mol. Pharmacol., 94, 2018
|
|
5E0G
 
 | | 1.20 A resolution structure of Norovirus 3CL protease in complex with a triazole-based macrocyclic (17-mer) inhibitor | | Descriptor: | (phenylmethyl) ~{N}-[(8~{S},11~{S},14~{S})-8-(hydroxymethyl)-11-(2-methylpropyl)-5,10,13-tris(oxidanylidene)-1,4,9,12,17,18-hexazabicyclo[14.2.1]nonadeca-16(19),17-dien-14-yl]carbamate, CHLORIDE ION, Norovirus 3C-like protease | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Weerawarna, P.M, Kim, Y, Kankanamalage, A.C.G, Damalanka, V.C, Lushington, G.H, Alliston, K.R, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2015-09-28 | | Release date: | 2016-05-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-based design and synthesis of triazole-based macrocyclic inhibitors of norovirus protease: Structural, biochemical, spectroscopic, and antiviral studies.
Eur.J.Med.Chem., 119, 2016
|
|
6PEK
 
 | | Structure of Spastin Hexamer (Subunit A-E) in complex with substrate peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Han, H, Schubert, H.L, McCullough, J, Monroe, N, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2019-06-20 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of spastin bound to a glutamate-rich peptide implies a hand-over-hand mechanism of substrate translocation.
J.Biol.Chem., 295, 2020
|
|
7CLF
 
 | | PigF with SAH | | Descriptor: | ACETATE ION, Methyltransferase domain-containing protein, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Qiu, S, Xu, D, Han, N, Sun, B, Ran, T, Wang, W. | | Deposit date: | 2020-07-20 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | Crystal structures of PigF, an O-methyltransferase involved in the prodigiosin synthetic pathway, reveal an induced-fit substrate-recognition mechanism.
Iucrj, 9, 2022
|
|
6FWE
 
 | | Phosphotriesterase PTE_C23_6 | | Descriptor: | 1-[methoxy(methyl)phosphoryl]oxyethane, 1-ethyl-1-methyl-cyclohexane, FORMIC ACID, ... | | Authors: | Dym, O, Aggarwal, N, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Leader, H, Ashani, Y, Goldsmith, M, Greisen, P, Tawfik, D, Sussman, L.J. | | Deposit date: | 2018-03-06 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.774 Å) | | Cite: | Phosphotriesterase
PTE_C23_6
To Be Published
|
|
5E4P
 
 | | X-ray Crystal Structure Analysis of Magnetically Oriented Microcrystals of Lysozyme at 1.8 angstrom Resolution | | Descriptor: | Lysozyme C | | Authors: | Tsukui, S, Kimura, F, Garman, E.F, Baba, S, Mizuno, N, Mikami, B, Kimura, T. | | Deposit date: | 2015-10-06 | | Release date: | 2016-08-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | X-ray crystal structure analysis of magnetically oriented microcrystals of lysozyme at 1.8 A resolution
J.Appl.Crystallogr., 49, 2016
|
|
7CLU
 
 | | PigF with SAH | | Descriptor: | ACETATE ION, GLYCEROL, Methyltransferase domain-containing protein | | Authors: | Qiu, S, Xu, D, Han, N, Sun, B, Ran, T, Wang, W. | | Deposit date: | 2020-07-22 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of PigF, an O-methyltransferase involved in the prodigiosin synthetic pathway, reveal an induced-fit substrate-recognition mechanism.
Iucrj, 9, 2022
|
|
1REM
 
 | | HUMAN LYSOZYME WITH MAN-B1,4-GLCNAC COVALENTLY ATTACHED TO ASP53 | | Descriptor: | LYSOZYME, NITRATE ION, R-1,2-PROPANEDIOL, ... | | Authors: | Muraki, M, Harata, K, Sugita, N, Sato, K. | | Deposit date: | 1998-01-14 | | Release date: | 1998-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structure of human lysozyme labelled with 2',3'-epoxypropyl beta-glycoside of man-beta1,4-GlcNAc. Structural change and recognition specificity at subsite B.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
6G87
 
 | | Flavonoid-responsive Regulator FrrA | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, TetR/AcrR family transcriptional regulator | | Authors: | Werner, N, Hoppen, J, Palm, G, Werten, S, Goettfert, M, Hinrichs, W. | | Deposit date: | 2018-04-07 | | Release date: | 2019-04-24 | | Last modified: | 2021-08-18 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | The induction mechanism of the flavonoid-responsive regulator FrrA.
Febs J., 2021
|
|
5E7T
 
 | | Structure of the tripod (BppUct-A-L) from the baseplate of bacteriophage Tuc2009 | | Descriptor: | CALCIUM ION, Major structural protein 1, Minor structural protein 4, ... | | Authors: | Legrand, P, Collins, B, Blangy, S, Murphy, J, Spinelli, S, Gutierrez, C, Richet, N, Kellenberger, C, Desmyter, A, Mahony, J, van Sinderen, D, Cambillau, C. | | Deposit date: | 2015-10-13 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Atomic Structure of the Phage Tuc2009 Baseplate Tripod Suggests that Host Recognition Involves Two Different Carbohydrate Binding Modules.
Mbio, 7, 2016
|
|
6DMQ
 
 | |
6G52
 
 | | CRYSTAL STRUCTURE OF THE CNMP BINDING DOMAIN OF THE MAGNESIUM TRANSPORTER CNNM4 | | Descriptor: | Metal transporter CNNM4 | | Authors: | Gimenez, P, Oyenarte, I, Hardy, S, Zubillaga, M, Merino, N, Blanco, F.J, Siliqi, D, Tremblay, M, Muller, D, Martinez-Cruz, L.A. | | Deposit date: | 2018-03-28 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.691 Å) | | Cite: | Structural Insights into the Intracellular Region of the Human Magnesium Transport Mediator CNNM4.
Int J Mol Sci, 20, 2019
|
|
6DO4
 
 | | KLHDC2 ubiquitin ligase in complex with SelS C-end degron | | Descriptor: | Kelch domain-containing protein 2, SELS C-END DEGRON | | Authors: | Rusnac, D.V, Lin, H.C, Yen, H.C.S, Zheng, N. | | Deposit date: | 2018-06-08 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Recognition of the Diglycine C-End Degron by CRL2KLHDC2Ubiquitin Ligase.
Mol. Cell, 72, 2018
|
|
5E7F
 
 | | Complex between lactococcal phage Tuc2009 RBP head domain and a nanobody (L06) | | Descriptor: | Major structural protein 1, nanobody L06 | | Authors: | Legrand, P, Collins, B, Blangy, S, Murphy, J, Spinelli, S, Gutierrez, C, Richet, N, Kellenberger, C, Desmyter, A, Mahony, J, van Sinderen, D, Cambillau, C. | | Deposit date: | 2015-10-12 | | Release date: | 2015-12-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Atomic Structure of the Phage Tuc2009 Baseplate Tripod Suggests that Host Recognition Involves Two Different Carbohydrate Binding Modules.
Mbio, 7, 2016
|
|
1RZO
 
 | | Agglutinin from Ricinus communis with galactoaza | | Descriptor: | Agglutinin, SULFATE ION, beta-D-galactopyranose | | Authors: | Gabdoulkhakov, A.G, Savochkina, Y, Konareva, N, Krauspenhaar, R, Stoeva, S, Nikonov, S.V, Voelter, W, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2003-12-26 | | Release date: | 2004-12-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structure-function investigation complex of Agglutinin from Ricinus communis with galactoaza
To be Published
|
|
6FO8
 
 | | Vitamin D nuclear receptor complex 4 | | Descriptor: | (1~{R},3~{S},5~{Z})-4-methylidene-5-[(~{E})-3-[3-(6-methyl-6-oxidanyl-heptyl)phenyl]non-2-enylidene]cyclohexane-1,3-diol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N. | | Deposit date: | 2018-02-06 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Aromatic-Based Design of Highly Active and Noncalcemic Vitamin D Receptor Agonists.
J. Med. Chem., 61, 2018
|
|
1S19
 
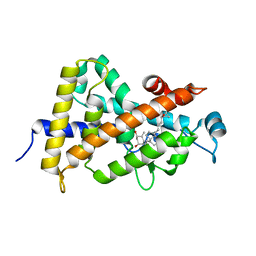 | | Crystal structure of VDR ligand binding domain complexed to calcipotriol. | | Descriptor: | CALCIPOTRIOL, Vitamin D3 receptor | | Authors: | Tocchini-Valentini, G, Rochel, N, Wurtz, J.M, Moras, D. | | Deposit date: | 2004-01-06 | | Release date: | 2004-04-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the vitamin D nuclear receptor liganded with the vitamin D side chain analogues calcipotriol and seocalcitol, receptor agonists of clinical importance. Insights into a structural basis for the switching of calcipotriol to a receptor antagonist by further side chain modification.
J.Med.Chem., 47, 2004
|
|
6CAN
 
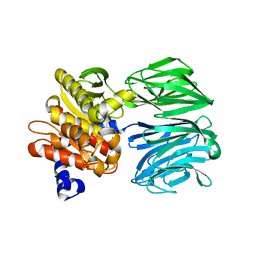 | |
6V5M
 
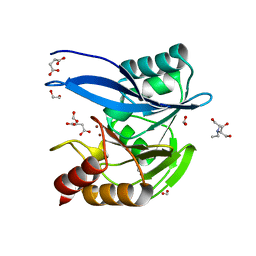 | | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica in Complex with Succinate | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase, ... | | Authors: | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-04 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica in Complex with Succinate.
To Be Published
|
|
1SH2
 
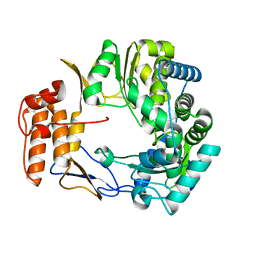 | | Crystal Structure of Norwalk Virus Polymerase (Metal-free, Centered Orthorhombic) | | Descriptor: | RNA Polymerase | | Authors: | Ng, K.K, Pendas-Franco, N, Rojo, J, Boga, J.A, Machin, A, Alonso, J.M, Parra, F. | | Deposit date: | 2004-02-24 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of norwalk virus polymerase reveals the carboxyl terminus in the active site cleft.
J.Biol.Chem., 279, 2004
|
|
6GB2
 
 | | Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM. This file contains the 39S ribosomal subunit. | | Descriptor: | 'Mitochondrial ribosomal protein L30, 'Mitochondrial ribosomal protein L55, 'Mitochondrial ribosomal protein L59, ... | | Authors: | Kummer, E, Leibundgut, M, Boehringer, D, Ban, N. | | Deposit date: | 2018-04-13 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM.
Nature, 560, 2018
|
|
6V6R
 
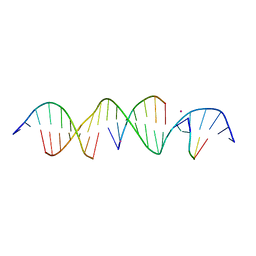 | | Crystal Structure of a Bromine Derivatized Self-Assembling DNA Crystal Scaffold with Rhombohedral Symmetry. | | Descriptor: | CACODYLATE ION, DNA (5'-D(*CP*AP*CP*TP*GP*AP*CP*TP*CP*AP*TP*GP*CP*TP*CP*AP*TP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*AP*GP*AP*TP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2019-12-05 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Self-Assembled Rhombohedral DNA Crystal Scaffold with Tunable Cavity Sizes and High-Resolution Structural Detail.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6V71
 
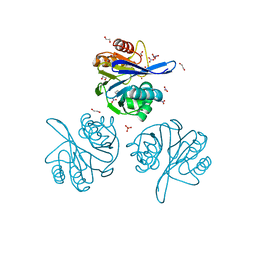 | | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica with Nitrate in the Active Site | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, FORMIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-06 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica with Nitrate in the Active Site
To Be Published
|
|
1SKG
 
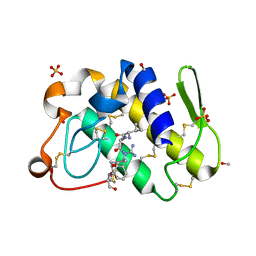 | | Structure-based rational drug design: Crystal structure of the complex formed between Phospholipase A2 and a pentapeptide Val-Ala-Phe-Arg-Ser | | Descriptor: | METHANOL, Phospholipase A2, SULFATE ION, ... | | Authors: | Ethayathulla, A.S, Singh, N, Sharma, S, Makker, J, Dey, S, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2004-03-04 | | Release date: | 2004-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structure-based rational drug design: Crystal structure of the complex formed between Phospholipase A2 and a pentapeptide Val-Ala-Phe-Arg-Ser
To be Published
|
|
