4M3D
 
 | | Rapid and efficient design of new inhibitors of Mycobacterium tuberculosis transcriptional repressor EthR using fragment growing, merging and linking approaches | | Descriptor: | 4-{3-[(phenylsulfonyl)amino]prop-1-yn-1-yl}-N-(3,3,3-trifluoropropyl)benzamide, HTH-type transcriptional regulator EthR | | Authors: | Villemagne, B, Flipo, M, Blondiaux, N, Crauste, C, Malaquin, S, Leroux, F, Piveteau, C, Villeret, V, Brodin, P, Villoutreix, B, Sperandio, O, Wohlkonig, A, Wintjens, R, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2013-08-06 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand Efficiency Driven Design of New Inhibitors of Mycobacterium tuberculosis Transcriptional Repressor EthR Using Fragment Growing, Merging, and Linking Approaches.
J.Med.Chem., 57, 2014
|
|
1WFS
 
 | | Solution Structure of Glia Maturation Factor-gamma from Mus Musculus | | Descriptor: | Glia maturation factor gamma | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-26 | | Release date: | 2004-11-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution structures of actin depolymerizing factor homology domains.
Protein Sci., 18, 2009
|
|
2L90
 
 | | Solution structure of murine myristoylated msrA | | Descriptor: | MYRISTIC ACID, Peptide methionine sulfoxide reductase | | Authors: | Gruschus, J.M, Lim, J, Piszczek, G, Levine, R.L, Tjandra, N. | | Deposit date: | 2011-01-27 | | Release date: | 2012-01-11 | | Last modified: | 2012-08-01 | | Method: | SOLUTION NMR | | Cite: | Characterization and solution structure of mouse myristoylated methionine sulfoxide reductase A.
J.Biol.Chem., 287, 2012
|
|
4M3E
 
 | | Rapid and efficient design of new inhibitors of Mycobacterium tuberculosis transcriptional repressor EthR using fragment growing, merging and linking approaches | | Descriptor: | 4-(2-{[(propylsulfonyl)amino]methyl}-1,3-thiazol-4-yl)-N-(3,3,3-trifluoropropyl)benzamide, HTH-type transcriptional regulator EthR | | Authors: | Villemagne, B, Flipo, M, Blondiaux, N, Crauste, C, Malaquin, S, Leroux, F, Piveteau, C, Villeret, V, Brodin, P, Villoutreix, B, Sperandio, O, Wohlkonig, A, Wintjens, R, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2013-08-06 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.109 Å) | | Cite: | Ligand Efficiency Driven Design of New Inhibitors of Mycobacterium tuberculosis Transcriptional Repressor EthR Using Fragment Growing, Merging, and Linking Approaches.
J.Med.Chem., 57, 2014
|
|
4M3G
 
 | | Rapid and efficient design of new inhibitors of Mycobacterium tuberculosis transcriptional repressor EthR using fragment growing, merging and linking approaches | | Descriptor: | 4-(2-methyl-1,3-thiazol-4-yl)-N-(3,3,3-trifluoropropyl)benzenesulfonamide, HTH-type transcriptional regulator EthR | | Authors: | Villemagne, B, Flipo, M, Blondiaux, N, Crauste, C, Malaquin, S, Leroux, F, Piveteau, C, Villeret, V, Brodin, P, Villoutreix, B, Sperandio, O, Wohlkonig, A, Wintjens, R, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2013-08-06 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ligand Efficiency Driven Design of New Inhibitors of Mycobacterium tuberculosis Transcriptional Repressor EthR Using Fragment Growing, Merging, and Linking Approaches.
J.Med.Chem., 57, 2014
|
|
3A5P
 
 | | Crystal structure of hemagglutinin | | Descriptor: | Haemagglutinin I | | Authors: | Watanabe, N, Sakai, N, Nakamura, T, Nabeshima, Y, Kouno, T, Mizuguchi, M, Kawano, K. | | Deposit date: | 2009-08-10 | | Release date: | 2010-08-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Structure of Physarum polycephalum hemagglutinin I suggests a minimal carbohydrate recognition domain of legume lectin fold
J.Mol.Biol., 405, 2011
|
|
1X67
 
 | | Solution structure of the cofilin homology domain of HIP-55 (drebrin-like protein) | | Descriptor: | Drebrin-like protein | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Sato, M, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-17 | | Release date: | 2005-11-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution structures of actin depolymerizing factor homology domains.
Protein Sci., 18, 2009
|
|
2KP3
 
 | | Structure of ANA-RNA hybrid duplex | | Descriptor: | RNA (5'-R(*(GAO)P*(CAR)P*(UAR)P*(A5O)P*(UAR)P*(A5O)P*(A5O)P*(UAR)P*(GAO)P*(GAO))-3'), RNA (5'-R(*CP*CP*AP*UP*UP*AP*UP*AP*GP*C)-3') | | Authors: | Gonzalez, C, Martn-Pintado, N, Watts, J, Gomez-Pinto, I, Dhama, M, Orozco, M, Schwartzentruber, J, Portella, G. | | Deposit date: | 2009-10-06 | | Release date: | 2010-02-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Differential stability of 2'F-ANA*RNA and ANA*RNA hybrid duplexes: roles of structure, pseudohydrogen bonding, hydration, ion uptake and flexibility.
Nucleic Acids Res., 38, 2010
|
|
2A85
 
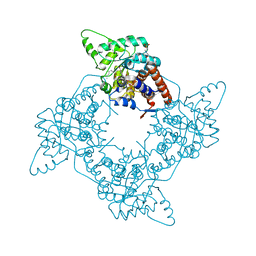 | | Crystal Structure of the G81A mutant of the Active Chimera of (S)-Mandelate Dehydrogenase in complex with its substrate 2-hydroxyoctanoate | | Descriptor: | (2S)-2-HYDROXYOCTANOIC ACID, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Sukumar, N, Xu, Y, Mitra, B, Mathews, F.S. | | Deposit date: | 2005-07-07 | | Release date: | 2006-07-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the G81A mutant form of the active chimera of (S)-mandelate dehydrogenase and its complex with two of its substrates.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2A7N
 
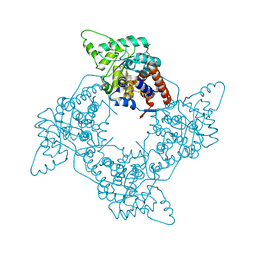 | | Crystal Structure of the G81A mutant of the Active Chimera of (S)-Mandelate Dehydrogenase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN MONONUCLEOTIDE, L(+)-mandelate dehydrogenase | | Authors: | Sukumar, N, Xu, Y, Mitra, B, Mathews, F.S. | | Deposit date: | 2005-07-05 | | Release date: | 2006-07-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the G81A mutant form of the active chimera of (S)-mandelate dehydrogenase and its complex with two of its substrates
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2LZ6
 
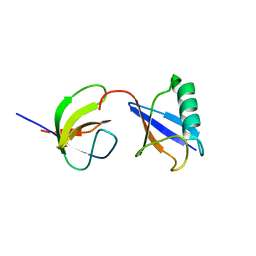 | | Distinct ubiquitin binding modes exhibited by sh3 domains: molecular determinants and functional implications | | Descriptor: | CD2-associated protein, Ubiquitin | | Authors: | Ortega-Roldan, J, Azuaga, A, Blackledge, M, Van Nuland, N. | | Deposit date: | 2012-09-24 | | Release date: | 2013-10-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Distinct Ubiquitin Binding Modes Exhibited by SH3 Domains: Molecular Determinants and Functional Implications.
Plos One, 8, 2013
|
|
2F8W
 
 | | Crystal structure of d(CACGTG)2 | | Descriptor: | 1,3-DIAMINOPROPANE, 5'-D(*CP*AP*CP*GP*TP*G)-3', SPERMINE | | Authors: | Narayana, N, Shamala, N, Ganesh, K.N, Viswamitra, M.A. | | Deposit date: | 2005-12-04 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Interaction between the Z-Type DNA Duplex and 1,3-Propanediamine: Crystal Structure of d(CACGTG)2 at 1.2 A Resolution
Biochemistry, 45, 2006
|
|
2PLY
 
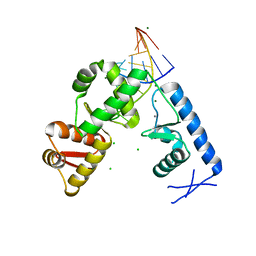 | | Structure of the mRNA binding fragment of elongation factor SelB in complex with SECIS RNA. | | Descriptor: | CALCIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Soler, N, Fourmy, D, Yoshizawa, S. | | Deposit date: | 2007-04-20 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insight into a molecular switch in tandem winged-helix motifs from elongation factor SelB.
J.Mol.Biol., 370, 2007
|
|
2PQW
 
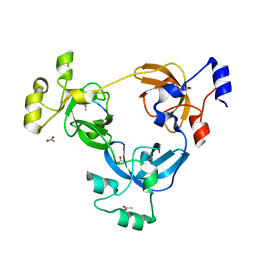 | | Crystal structure of L3MBTL1 in complex with H4K20Me2 (residues 17-25), trigonal form | | Descriptor: | ACETATE ION, Histone H4, Lethal(3)malignant brain tumor-like protein | | Authors: | Allali-Hassani, A, Liu, Y, Herzanych, N, Ouyang, H, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-02 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | L3MBTL1 recognition of mono- and dimethylated histones.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2PIK
 
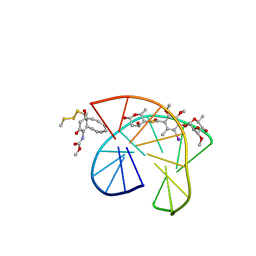 | | CALICHEAMICIN GAMMA1I-DNA COMPLEX, NMR, 6 STRUCTURES | | Descriptor: | 2,4-dideoxy-4-(ethylamino)-3-O-methyl-alpha-L-threo-pentopyranose-(1-2)-4-amino-4,6-dideoxy-beta-D-glucopyranose, 2,6-dideoxy-4-thio-beta-D-allopyranose, 3-O-methyl-alpha-L-rhamnopyranose, ... | | Authors: | Kumar, R.A, Ikemoto, N, Patel, D.J. | | Deposit date: | 1996-12-31 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the calicheamicin gamma 1I-DNA complex.
J.Mol.Biol., 265, 1997
|
|
2MQI
 
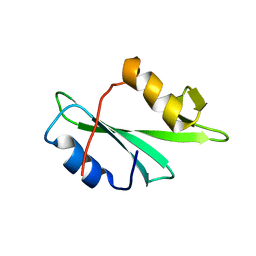 | |
2PJP
 
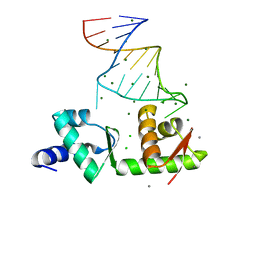 | | Structure of the mRNA-binding domain of elongation factor SelB from E.coli in complex with SECIS RNA | | Descriptor: | CALCIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Soler, N, Fourmy, D, Yoshizawa, S. | | Deposit date: | 2007-04-16 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into a molecular switch in tandem winged-helix motifs from elongation factor SelB.
J.Mol.Biol., 370, 2007
|
|
7RQD
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, A-site deacylated tRNA analog CACCA, P-site MTI-tripeptidyl-tRNA analog ACCA-ITM, and chloramphenicol at 2.50A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Syroegin, E.A, Flemmich, L, Klepacki, D, Vazquez-Laslop, N, Micura, R, Polikanov, Y.S. | | Deposit date: | 2021-08-06 | | Release date: | 2022-01-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the context-specific action of the classic peptidyl transferase inhibitor chloramphenicol.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7LTN
 
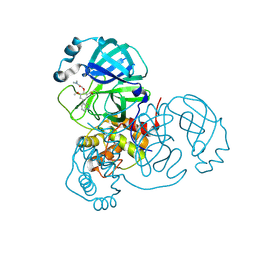 | | Crystal structure of Mpro in complex with inhibitor CDD-1713 | | Descriptor: | 2-[4-(1~{H}-indazol-4-yl)-2-methanoyl-6-methoxy-phenoxy]-~{N},~{N}-dimethyl-ethanamide, 3C-like proteinase | | Authors: | Lu, S, Palzkill, T, Matzuk, M, Young, D, Melek, N, Chamakuri, S. | | Deposit date: | 2021-02-19 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | DNA-encoded chemistry technology yields expedient access to SARS-CoV-2 M pro inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MTG
 
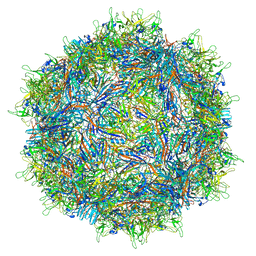 | | Structure of the adeno-associated virus 9 capsid at pH 6.0 | | Descriptor: | Capsid protein VP1 | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
7RKX
 
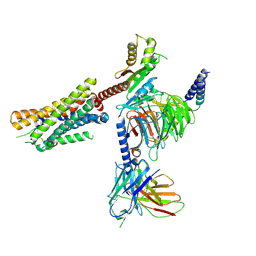 | | Structure of US27-Gi-scFv16 in CL-state | | Descriptor: | Antibody fragment scFv16, G-protein coupled receptor homolog US27, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Tsutsumi, N, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-07-22 | | Release date: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Atypical structural snapshots of human cytomegalovirus GPCR interactions with host G proteins
Sci Adv, 8, 2022
|
|
7RQE
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, A-site deacylated tRNA analog CACCA, P-site MAI-tripeptidyl-tRNA analog ACCA-IAM, and chloramphenicol at 2.40A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Syroegin, E.A, Flemmich, L, Klepacki, D, Vazquez-Laslop, N, Micura, R, Polikanov, Y.S. | | Deposit date: | 2021-08-06 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the context-specific action of the classic peptidyl transferase inhibitor chloramphenicol.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7MTP
 
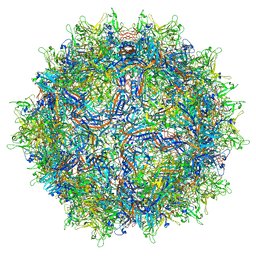 | | Structure of the adeno-associated virus 9 capsid at pH 5.5 | | Descriptor: | Capsid protein VP1 | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
7MTW
 
 | | Structure of the adeno-associated virus 9 capsid at pH 4.0 | | Descriptor: | Capsid protein VP1 | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
5C4L
 
 | | Conformational alternate of sisomicin in complex with APH(2")-IVa | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, (2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-ol, APH(2'')-Id | | Authors: | Kaplan, E, Guichou, J.F, Berrou, K, Chaloin, L, Leban, N, Lallemand, P, Barman, T, Serpersu, E.H, Lionne, C. | | Deposit date: | 2015-06-18 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Aminoglycoside binding and catalysis specificity of aminoglycoside 2-phosphotransferase IVa: A thermodynamic, structural and kinetic study.
Biochim.Biophys.Acta, 1860, 2016
|
|
