8WA2
 
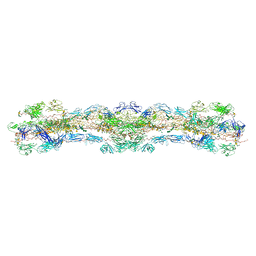 | | cryo-EM structure of native mastigonemes isolated from Chlamydomonas reinhardtii at 3.0 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Huang, J, Tao, H, Chen, J, Pan, J, Yan, C, Yan, N. | | Deposit date: | 2023-09-06 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure-guided discovery of protein and glycan components in native mastigonemes.
Cell, 187, 2024
|
|
6SHI
 
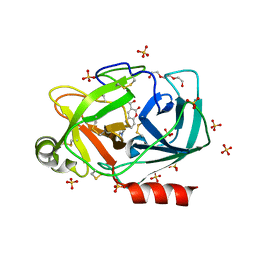 | |
8WCQ
 
 | | Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in intermediate state | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHLORIDE ION, Proton-gated ion channel | | Authors: | Bharambe, N, Li, Z, Basak, S. | | Deposit date: | 2023-09-13 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cryo-EM structures of prokaryotic ligand-gated ion channel GLIC provide insights into gating in a lipid environment.
Nat Commun, 15, 2024
|
|
6D2X
 
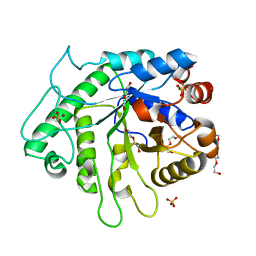 | | Crystal structure of the GH26 domain from PbGH26-GH5A endo-beta-mannanase/endo-beta-glucanase from Prevotella bryantii | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Aryl-phospho-beta-D-glucosidase BglC, GH1 family, ... | | Authors: | Stogios, P.J, Skarina, T, McGregor, N, Di Leo, R, Brumer, H, Savchenko, A. | | Deposit date: | 2018-04-14 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of the GH26 domain from PbGH26-GH5A endo-beta-mannanase/endo-beta-glucanase from Prevotella bryantii
To Be Published
|
|
8W15
 
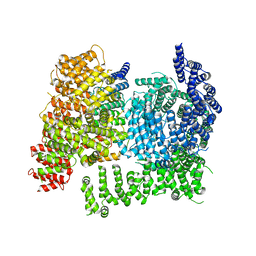 | |
6CMC
 
 | |
2MWQ
 
 | | Solution structure of PsbQ from spinacia oleracea | | Descriptor: | Oxygen-evolving enhancer protein 3, chloroplastic | | Authors: | Rathner, P, Mueller, N, Wimmer, R, Chandra, K. | | Deposit date: | 2014-11-19 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR and molecular dynamics reveal a persistent alpha helix within the dynamic region of PsbQ from photosystem II of higher plants.
Proteins, 83, 2015
|
|
7JXC
 
 | | Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology | | Descriptor: | NONAETHYLENE GLYCOL, S2H14 antigen-binding (Fab) fragment | | Authors: | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-27 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
8WCR
 
 | | Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in open state | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHLORIDE ION, Proton-gated ion channel | | Authors: | Bharambe, N, Li, Z, Basak, S. | | Deposit date: | 2023-09-13 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Cryo-EM structures of prokaryotic ligand-gated ion channel GLIC provide insights into gating in a lipid environment.
Nat Commun, 15, 2024
|
|
6SHJ
 
 | | Escherichia coli AGPase in complex with FBP. Symmetry applied C2 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Glucose-1-phosphate adenylyltransferase | | Authors: | Cifuente, J.O, Comino, N, D'Angelo, C, Marina, A, Gil-Carton, D, Albesa-Jove, D, Guerin, M.E. | | Deposit date: | 2019-08-07 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The allosteric control mechanism of bacterial glycogen biosynthesis disclosed by cryoEM.
Curr Res Struct Biol, 2, 2020
|
|
6CIH
 
 | | Crystal structure of a group II intron lariat in the post-catalytic state | | Descriptor: | IRIDIUM HEXAMMINE ION, MAGNESIUM ION, RNA (5'-R(P*UP*GP*UP*UP*UP*AP*UP*UP*AP*AP*AP*AP*AP*C*-3'), ... | | Authors: | Chan, R.T, Peters, J.K, Robart, A.R, Wiryaman, T, Rajashankar, K.R, Toor, N. | | Deposit date: | 2018-02-23 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.676 Å) | | Cite: | Structural basis for the second step of group II intron splicing.
Nat Commun, 9, 2018
|
|
6CRX
 
 | | SARS Spike Glycoprotein, Stabilized variant, two S1 CTDs in the upwards conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
6CUL
 
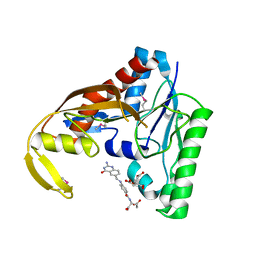 | | PvdF of pyoverdin biosynthesis is a structurally unique N10-formyltetrahydrofolate-dependent formyltransferase | | Descriptor: | CITRIC ACID, N-(4-{[(2-amino-4-oxo-1,4-dihydroquinazolin-6-yl)methyl]amino}benzene-1-carbonyl)-D-glutamic acid, Pyoverdine synthetase F | | Authors: | Kenjic, N, Hoag, M.R, Moraski, G.C, Caperelli, C.A, Moran, G.R, Lamb, A.L. | | Deposit date: | 2018-03-26 | | Release date: | 2019-02-06 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | PvdF of pyoverdin biosynthesis is a structurally unique N10-formyltetrahydrofolate-dependent formyltransferase.
Arch. Biochem. Biophys., 664, 2019
|
|
8VI4
 
 | | TehA from Haemophilus influenzae purified in LMNG | | Descriptor: | Tellurite resistance protein TehA homolog | | Authors: | Catalano, C, Senko, S, Tran, N.L, Lucier, K.W, Farwell, A.C, Silva, M.S, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | High-Resolution Cryo-Electron Microscopy Structure Determination of Haemophilus influenzae Tellurite-Resistance Protein A via 200 kV Transmission Electron Microscopy.
Int J Mol Sci, 25, 2024
|
|
8VSP
 
 | | Cryo-EM structure of human invariant chain in complex with HLA-DQ | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen gamma chain, HLA class II histocompatibility antigen, ... | | Authors: | Wang, N, Caveney, N.A, Jude, K.M, Garcia, K.C. | | Deposit date: | 2024-01-24 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structural insights into human MHC-II association with invariant chain.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6S8E
 
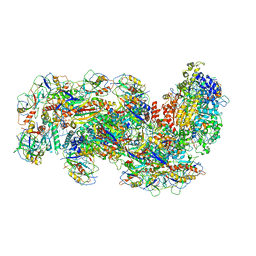 | | Cryo-EM structure of the type III-B Cmr-beta complex bound to non-cognate target RNA | | Descriptor: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | Authors: | Sofos, N, Montoya, G, Stella, S. | | Deposit date: | 2019-07-09 | | Release date: | 2020-07-08 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
8VI5
 
 | | TehA from Haemophilus influenzae purified in OG | | Descriptor: | Tellurite resistance protein TehA homolog | | Authors: | Catalano, C, Senko, S, Tran, N.L, Lucier, K.W, Farwell, A.C, Silva, M.S, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-Resolution Cryo-Electron Microscopy Structure Determination of Haemophilus influenzae Tellurite-Resistance Protein A via 200 kV Transmission Electron Microscopy.
Int J Mol Sci, 25, 2024
|
|
8VI3
 
 | | TehA from Haemophilus influenzae purified in GDN | | Descriptor: | Tellurite resistance protein TehA homolog | | Authors: | Catalano, C, Senko, S, Tran, N.L, Lucier, K.W, Farwell, A.C, Silva, M.S, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | High-Resolution Cryo-Electron Microscopy Structure Determination of Haemophilus influenzae Tellurite-Resistance Protein A via 200 kV Transmission Electron Microscopy.
Int J Mol Sci, 25, 2024
|
|
8VI2
 
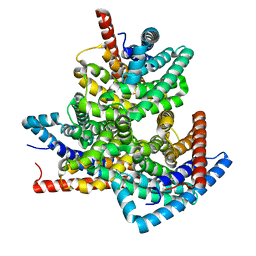 | | TehA from Haemophilus influenzae purified in DDM | | Descriptor: | Tellurite resistance protein TehA homolog | | Authors: | Catalano, C, Senko, S, Tran, N.L, Lucier, K.W, Farwell, A.C, Silva, M.S, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | High-Resolution Cryo-Electron Microscopy Structure Determination of Haemophilus influenzae Tellurite-Resistance Protein A via 200 kV Transmission Electron Microscopy.
Int J Mol Sci, 25, 2024
|
|
6SH8
 
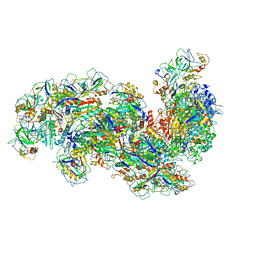 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA and AMPPnP, state 2, in the presence of ssDNA | | Descriptor: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | Authors: | Sofos, N, Montoya, G, Stella, S. | | Deposit date: | 2019-08-06 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
8VRW
 
 | | Cryo-EM structure of human invariant chain in complex with HLA-DR15 | | Descriptor: | HLA class II histocompatibility antigen gamma chain, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Wang, N, Caveney, N.A, Jude, K.M, Garcia, K.C. | | Deposit date: | 2024-01-22 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural insights into human MHC-II association with invariant chain.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8W01
 
 | |
6SMT
 
 | | S-enantioselective imine reductase from Mycobacterium smegmatis | | Descriptor: | (2S)-2-ethylhexan-1-ol, 1,2-ETHANEDIOL, 6-phosphogluconate dehydrogenase, ... | | Authors: | Meyer, T, Zumbraegel, N, Geerds, C, Groeger, H, Niemann, H.H. | | Deposit date: | 2019-08-22 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Characterization of an S -enantioselective Imine Reductase from Mycobacterium Smegmatis .
Biomolecules, 10, 2020
|
|
7JPE
 
 | | Room Temperature Structure of SARS-CoV-2 Nsp10/Nsp16 Methyltransferase in a Complex with m7GpppA Cap-0 and SAM Determined by Fixed-Target Serial Crystallography | | Descriptor: | 2'-O-methyltransferase, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Non-structural protein 10, ... | | Authors: | Wilamowski, M, Sherrell, D.A, Minasov, G, Kim, Y, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-08-07 | | Release date: | 2020-08-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7JXE
 
 | | Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology | | Descriptor: | S2X35 antigen-binding (Fab) fragment | | Authors: | Tortorici, M.A, Park, Y.J, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-27 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.043 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
