7R36
 
 | | Difference-refined structure of fatty acid photodecarboxylase 2 microsecond following 400-nm laser irradiation of the dark-state determined by SFX | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | 著者 | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, De Zitter, E, Schlichting, I, Colletier, J.P, Weik, M. | | 登録日 | 2022-02-06 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Time-resolved serial femtosecond crystallography on fatty-acid photodecarboxylase: lessons learned.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7R33
 
 | | Difference-refined structure of fatty acid photodecarboxylase 20 ps following 400-nm laser irradiation of the dark-state determined by SFX | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | 著者 | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, De Zitter, E, Schlichting, I, Colletier, J.P, Weik, M. | | 登録日 | 2022-02-06 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Time-resolved serial femtosecond crystallography on fatty-acid photodecarboxylase: lessons learned.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7R34
 
 | | Difference-refined structure of fatty acid photodecarboxylase 900 ps following 400-nm laser irradiation of the dark-state determined by SFX | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | 著者 | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, De Zitter, E, Schlichting, I, Colletier, J.P, Weik, M. | | 登録日 | 2022-02-06 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Time-resolved serial femtosecond crystallography on fatty-acid photodecarboxylase: lessons learned.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7R35
 
 | | Difference-refined structure of fatty acid photodecarboxylase 300 ns following 400-nm laser irradiation of the dark-state determined by SFX | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | 著者 | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, De Zitter, E, Schlichting, I, Colletier, J.P, Weik, W. | | 登録日 | 2022-02-06 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Time-resolved serial femtosecond crystallography on fatty-acid photodecarboxylase: lessons learned.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
4ZRC
 
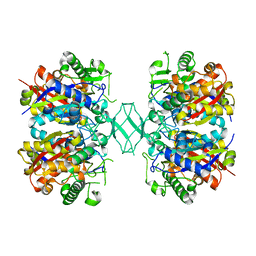 | | Crystal structure of MSM-13, a putative T1-like thiolase from Mycobacterium smegmatis | | 分子名称: | Beta-ketothiolase | | 著者 | Janardan, N, Harijan, R.K, Keima, T.R, Wierenga, R, Murthy, M.R.N. | | 登録日 | 2015-05-12 | | 公開日 | 2016-05-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural characterization of a mitochondrial 3-ketoacyl-CoA (T1)-like thiolase from Mycobacterium smegmatis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4YZF
 
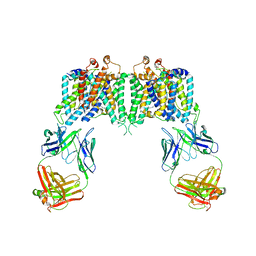 | | Crystal structure of the anion exchanger domain of human erythrocyte Band 3 | | 分子名称: | 2,2'-ethane-1,2-diylbis{5-[(sulfanylmethyl)amino]benzenesulfonic acid}, Band 3 anion transport protein, FAB fragment of Immunoglobulin (IgG) molecule | | 著者 | Alguel, Y, Arakawa, T, Yugiri, T.K, Iwanari, H, Hatae, H, Iwata, M, Abe, Y, Hino, T, Suno, C.I, Kuma, H, Kang, D, Murata, T, Hamakubo, T, Cameron, A.D, Kobayashi, T, Hamasaki, N, Iwata, S. | | 登録日 | 2015-03-25 | | 公開日 | 2015-11-04 | | 最終更新日 | 2015-11-18 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Crystal structure of the anion exchanger domain of human erythrocyte band 3.
Science, 350, 2015
|
|
7R0G
 
 | |
7EET
 
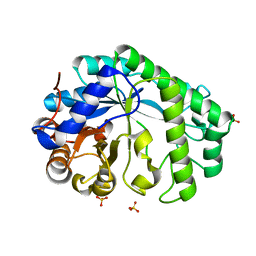 | | Mannanase KMAN from Klebsiella oxytoca KUB-CW2-3 | | 分子名称: | Mannanase KMAN, SULFATE ION | | 著者 | Pongsapipatana, N, Charoenwattanasatien, R, Pramanpol, N, Nitisinprasert, S, Keawsompong, S. | | 登録日 | 2021-03-19 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | Crystallization, structural characterization and kinetic analysis of a GH26 beta-mannanase from Klebsiella oxytoca KUB-CW2-3.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
8QZK
 
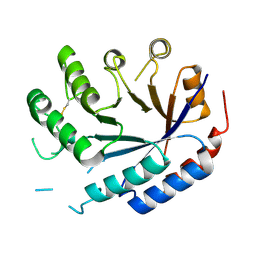 | | Catalytic core of endo-alpha-N-acetylgalactosaminidase from Bifidobacterium longum (EngBF) concieved by deep network hallucination: dEngBF4 Hexagonal form | | 分子名称: | ENDO-ALPHA-N-ACETYLGALACTOSAMINIDASE | | 著者 | Aghajari, N, Hansen, A.L, Thiesen, F.F, Crehuet, R, Marcos, E, Willemoes, M. | | 登録日 | 2023-10-27 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Carving out a Glycoside Hydrolase Active Site for Incorporation into a New Protein Scaffold Using Deep Network Hallucination.
Acs Synth Biol, 13, 2024
|
|
8PM8
 
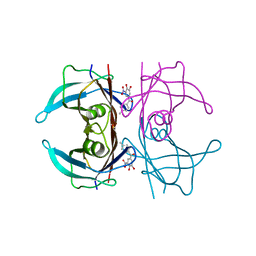 | | V30M Transthyretin structure in complex with Tolcalpone | | 分子名称: | Tolcapone, Transthyretin | | 著者 | Varejao, N, Pinheiro, F, Pallares, I, Ventura, S, Reverter, D. | | 登録日 | 2023-06-28 | | 公開日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | PITB: A high affinity transthyretin aggregation inhibitor with optimal pharmacokinetic properties.
Eur.J.Med.Chem., 261, 2023
|
|
8PM9
 
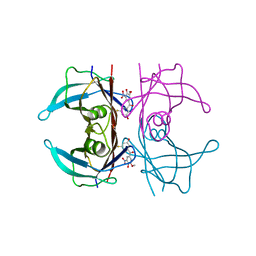 | | Crystal structure of human wild type transthyretin in complex with PITB (Pharmacokinetically Improved TTR Binder) | | 分子名称: | (3-fluoranyl-5-oxidanyl-phenyl)-(3-methoxy-5-nitro-4-oxidanyl-phenyl)methanone, Transthyretin | | 著者 | Varejao, N, Pinheiro, F, Pallares, I, Ventura, S, Reverter, D. | | 登録日 | 2023-06-28 | | 公開日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | PITB: A high affinity transthyretin aggregation inhibitor with optimal pharmacokinetic properties.
Eur.J.Med.Chem., 261, 2023
|
|
8PZ8
 
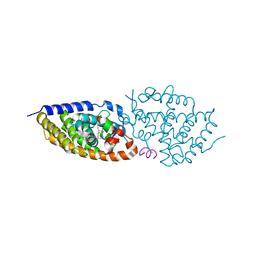 | | crystal structure of VDR in complex with D-Bishomo-1a,25-dihydroxyvitamin D3 Analog 54 | | 分子名称: | (1~{R},3~{R})-5-[(2~{E})-2-[(4~{a}~{R},5~{S},9~{a}~{S})-4~{a}-methyl-5-[(2~{R})-6-methyl-6-oxidanyl-heptan-2-yl]-3,4,5,6,7,8,9,9~{a}-octahydro-2~{H}-benzo[7]annulen-1-ylidene]ethylidene]cyclohexane-1,3-diol, Nuclear receptor coactivator 2, Vitamin D3 receptor A | | 著者 | Rochel, N. | | 登録日 | 2023-07-27 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.64 Å) | | 主引用文献 | Design, synthesis, and biological activity of D-bishomo-1 alpha ,25-dihydroxyvitamin D 3 analogs and their crystal structures with the vitamin D nuclear receptor.
Eur.J.Med.Chem., 271, 2024
|
|
4Z66
 
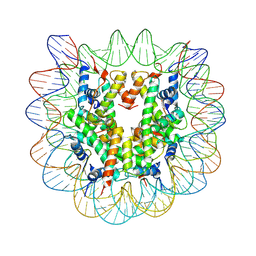 | | Nucleosome disassembly by RSC and SWI/SNF is enhanced by H3 acetylation near the nucleosome dyad axis | | 分子名称: | DNA (147-MER), Histone H2A, Histone H2B 1.1, ... | | 著者 | Dechassa, M.L, Luger, K, Chatterjee, N, North, J.A, Manohar, M, Prasad, R, Ottessen, J.J, Poirier, M.G, Bartholomew, B. | | 登録日 | 2015-04-03 | | 公開日 | 2015-10-14 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Histone Acetylation near the Nucleosome Dyad Axis Enhances Nucleosome Disassembly by RSC and SWI/SNF.
Mol.Cell.Biol., 35, 2015
|
|
8PZ6
 
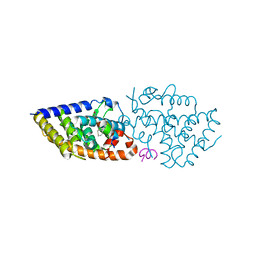 | | crystal structure of VDR in complex with D-Bishomo-1a,25-dihydroxyvitamin D3 analog 56 | | 分子名称: | (1~{R},3~{R})-5-[(2~{E})-2-[(4~{a}~{R},5~{S},9~{a}~{S})-4~{a}-methyl-5-[(2~{R})-6-methyl-6-oxidanyl-heptan-2-yl]-3,4,5,6,7,8,9,9~{a}-octahydro-2~{H}-benzo[7]annulen-1-ylidene]ethylidene]-2-(3-oxidanylpropylidene)cyclohexane-1,3-diol, Nuclear receptor coactivator 2, Vitamin D3 receptor A | | 著者 | Rochel, N. | | 登録日 | 2023-07-27 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Design, synthesis, and biological activity of D-bishomo-1 alpha ,25-dihydroxyvitamin D 3 analogs and their crystal structures with the vitamin D nuclear receptor.
Eur.J.Med.Chem., 271, 2024
|
|
4Z8J
 
 | |
7R3W
 
 | |
7RKL
 
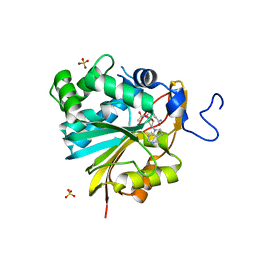 | | Structure of Nicotinamide N-Methyltransferase (NNMT) in complex with II399 (P1 space group) | | 分子名称: | 3-[3-(acetyl{[(1R,2R,3S,4R)-4-(4-chloro-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-2,3-dihydroxycyclopentyl]methyl}amino)prop-1-yn-1-yl]benzamide, NNMT protein, SULFATE ION | | 著者 | Yadav, R, Noinaj, N, Iyamu, I.D, Huang, R. | | 登録日 | 2021-07-22 | | 公開日 | 2022-07-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Exploring Unconventional SAM Analogues To Build Cell-Potent Bisubstrate Inhibitors for Nicotinamide N-Methyltransferase.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
6GPM
 
 | | Crystal structure of domain 2 from TmArgBP | | 分子名称: | Amino acid ABC transporter, periplasmic amino acid-binding protein | | 著者 | Smaldone, G, Balasco, N, Ruggiero, A, Berisio, R, Vitagliano, L. | | 登録日 | 2018-06-06 | | 公開日 | 2018-08-15 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Domain communication in Thermotoga maritima Arginine Binding Protein unraveled through protein dissection.
Int. J. Biol. Macromol., 119, 2018
|
|
4ZA1
 
 | | Crystal Structure of NosA Involved in Nosiheptide Biosynthesis | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, NosA | | 著者 | Liu, S, Guo, H, Zhang, T, Han, L, Yao, P, Zhang, Y, Rong, N, Yu, Y, Lan, W, Wang, C, Ding, J, Wang, R, Liu, W, Cao, C. | | 登録日 | 2015-04-13 | | 公開日 | 2015-08-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure-based Mechanistic Insights into Terminal Amide Synthase in Nosiheptide-Represented Thiopeptides Biosynthesis
Sci Rep, 5, 2015
|
|
8J4F
 
 | | Structure of human Nav1.7 in complex with Hardwickii acid | | 分子名称: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, (4~{a}~{R},5~{S},6~{R},8~{a}~{R})-5-[2-(furan-3-yl)ethyl]-5,6,8~{a}-trimethyl-3,4,4~{a},6,7,8-hexahydronaphthalene-1-carboxylic acid, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | 著者 | Wu, Q.R, Yan, N. | | 登録日 | 2023-04-19 | | 公開日 | 2023-06-14 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
7RKK
 
 | | Structure of Nicotinamide N-Methyltransferase (NNMT) in complex with II399 (C2 space group) | | 分子名称: | 3-[3-(acetyl{[(1R,2R,3S,4R)-4-(4-chloro-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-2,3-dihydroxycyclopentyl]methyl}amino)prop-1-yn-1-yl]benzamide, NNMT protein | | 著者 | Yadav, R, Noinaj, N, Iyamu, I.D, Huang, R. | | 登録日 | 2021-07-22 | | 公開日 | 2022-07-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.76 Å) | | 主引用文献 | Exploring Unconventional SAM Analogues To Build Cell-Potent Bisubstrate Inhibitors for Nicotinamide N-Methyltransferase.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
6GQD
 
 | | Structure of human galactose-1-phosphate uridylyltransferase (GALT), with crystallization epitope mutations A21Y:A22T:T23P:R25L | | 分子名称: | 1,2-ETHANEDIOL, 5,6-DIHYDROURIDINE-5'-MONOPHOSPHATE, Galactose-1-phosphate uridylyltransferase, ... | | 著者 | Fairhead, M, Strain-Damerell, C, Kopec, J, Bezerra, G.A, Zhang, M, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W, Structural Genomics Consortium (SGC) | | 登録日 | 2018-06-07 | | 公開日 | 2018-07-18 | | 実験手法 | X-RAY DIFFRACTION (1.523 Å) | | 主引用文献 | Structure of human galactose-1-phosphate uridylyltransferase (GALT), with crystallization epitope mutations A21Y:A22T:T23P:R25L
To Be Published
|
|
7RDN
 
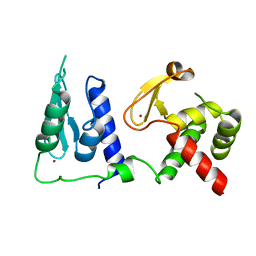 | | Crystal structure of S. cerevisiae pre-mRNA leakage protein 39 (Pml39) | | 分子名称: | Pre-mRNA leakage protein 39, ZINC ION | | 著者 | Hashimoto, H, Ramirez, D.H, Pawlak, N, Blobel, G, Palancade, B, Debler, E.W. | | 登録日 | 2021-07-09 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Structure of the pre-mRNA leakage 39-kDa protein reveals a single domain of integrated zf-C3HC and Rsm1 modules.
Sci Rep, 12, 2022
|
|
4ZDK
 
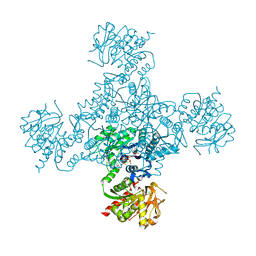 | | Crystal structure of the M. tuberculosis CTP synthase PyrG in complex with UTP, AMP-PCP and oxonorleucine | | 分子名称: | 5-OXO-L-NORLEUCINE, CTP synthase, MAGNESIUM ION, ... | | 著者 | Bellinzoni, M, Barilone, N, Alzari, P.M. | | 登録日 | 2015-04-17 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.49 Å) | | 主引用文献 | Thiophenecarboxamide Derivatives Activated by EthA Kill Mycobacterium tuberculosis by Inhibiting the CTP Synthetase PyrG.
Chem.Biol., 22, 2015
|
|
6GSA
 
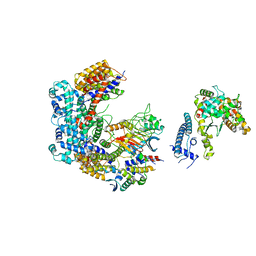 | | Core Centromere Binding Factor 3 (CBF3) with monomeric Ndc10 | | 分子名称: | Centromere DNA-binding protein complex CBF3 subunit A, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | 著者 | Zhang, W.J, Lukoynova, N, Vaughan, C.K. | | 登録日 | 2018-06-13 | | 公開日 | 2018-08-01 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Insights into Centromere DNA Bending Revealed by the Cryo-EM Structure of the Core Centromere Binding Factor 3 with Ndc10.
Cell Rep, 24, 2018
|
|
