6RIV
 
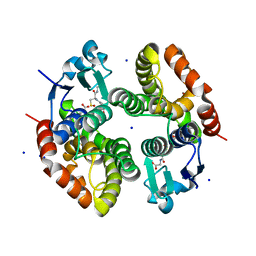 | | Crystal structure of Alopecurus myosuroides GSTF | | Descriptor: | GLYCEROL, Glutathione transferase, S-Hydroxy-Glutathione, ... | | Authors: | Papageorgiou, A.C, Poudel, N. | | Deposit date: | 2019-04-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Comparative structural and functional analysis of phi class glutathione transferases involved in multiple-herbicide resistance of grass weeds and crops.
Plant Physiol Biochem., 149, 2020
|
|
6RMD
 
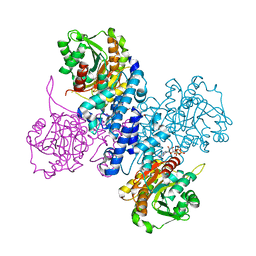 | | Structure of ATP bound Plasmodium falciparum IMP-nucleotidase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, IMP-specific 5'-nucleotidase, ... | | Authors: | Carrique, L, Ballut, L, Violot, S, Aghajari, N. | | Deposit date: | 2019-05-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and catalytic regulation of Plasmodium falciparum IMP specific nucleotidase.
Nat Commun, 11, 2020
|
|
6JQN
 
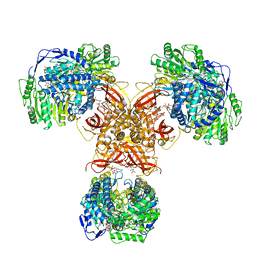 | | Structure of PaaZ, a bifunctional enzyme in complex with NADP+ and OCoA | | Descriptor: | Bifunctional protein PaaZ, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OCTANOYL-COENZYME A | | Authors: | Gakher, L, Vinothkumar, K.R, Katagihallimath, N, Sowdhamini, R, Sathyanarayanan, N, Cannone, G. | | Deposit date: | 2019-03-31 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway.
Nat Commun, 10, 2019
|
|
6JQO
 
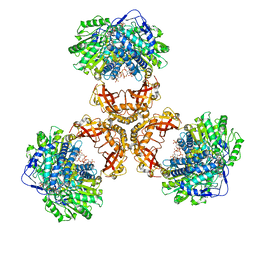 | | Structure of PaaZ, a bifunctional enzyme in complex with NADP+ and CCoA | | Descriptor: | Bifunctional protein PaaZ, CROTONYL COENZYME A, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Gakher, L, Vinothkumar, K.R, Katagihallimath, N, Sowdhamini, R, Sathyanarayanan, N, Cannone, G. | | Deposit date: | 2019-03-31 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway.
Nat Commun, 10, 2019
|
|
6UWN
 
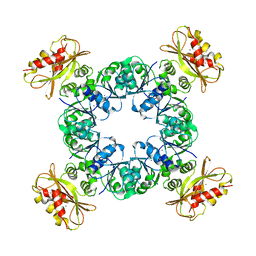 | |
6UX7
 
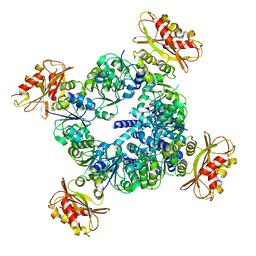 | |
6JQL
 
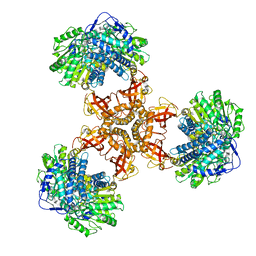 | | Structure of PaaZ, a bifunctional enzyme | | Descriptor: | Bifunctional protein PaaZ | | Authors: | Gakher, L, Vinothkumar, K.R, Katagihallimath, N, Sowdhamini, R, Sathyanarayanan, N, Cannone, G. | | Deposit date: | 2019-03-31 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway.
Nat Commun, 10, 2019
|
|
6JSF
 
 | | Crystal Structure of BACE1 in complex with N-(3-((4S,5S)-2-amino-4-methyl-5-phenyl-5,6-dihydro-4H-1,3-thiazin-4-yl)-4-fluorophenyl)-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, IODIDE ION, ... | | Authors: | Fujimoto, K, Matsuoka, E, Asada, N, Tadano, G, Yamamoto, T, Nakahara, K, Fuchino, K, Ito, H, Kanegawa, N, Moechars, D, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Selective beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors: Targeting the Flap to Gain Selectivity over BACE2.
J.Med.Chem., 62, 2019
|
|
6JQM
 
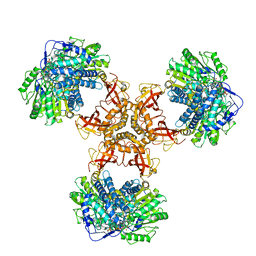 | | Structure of PaaZ with NADPH | | Descriptor: | Bifunctional protein PaaZ, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Gakher, L, Vinothkumar, K.R, Katagihallimath, N, Sowdhamini, R, Sathyanarayanan, N, Cannone, G. | | Deposit date: | 2019-03-31 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway.
Nat Commun, 10, 2019
|
|
6JXR
 
 | | Structure of human T cell receptor-CD3 complex | | Descriptor: | T cell receptor alpha variable 12-3,Possible J 11 gene segment,T cell receptor alpha constant, T cell receptor beta variable 6-5,M1-specific T cell receptor beta chain,T cell receptor beta constant 2, T-cell surface glycoprotein CD3 delta chain, ... | | Authors: | Dong, D, Zheng, L, Lin, J, Zhu, Y, Li, N, Zhang, B, Xie, S, Zheng, J, Wang, Y, Gao, N, Huang, Z. | | Deposit date: | 2019-04-24 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of assembly of the human T cell receptor-CD3 complex.
Nature, 573, 2019
|
|
6JT9
 
 | | Crystal Structure of D464A mutant of FGAM Synthetase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sharma, N, Ahalawat, N, Sandhu, P, Mondal, J, Anand, R. | | Deposit date: | 2019-04-10 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Role of allosteric switches and adaptor domains in long-distance cross-talk and transient tunnel formation.
Sci Adv, 6, 2020
|
|
6UX4
 
 | |
6SGX
 
 | | Structure of protomer 1 of the ESX-3 core complex | | Descriptor: | ESX-3 secretion system EccB3, ESX-3 secretion system protein EccC3, ESX-3 secretion system protein EccD3, ... | | Authors: | Famelis, N, Rivera-Calzada, A, Llorca, O, Geibel, S. | | Deposit date: | 2019-08-05 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Architecture of the mycobacterial type VII secretion system.
Nature, 576, 2019
|
|
6JSZ
 
 | | BACE2 xaperone complex with N-{3-[(5R)-3-amino-5-methyl-9,9-dioxo-2,9lambda6-dithia-4-azaspiro[5.5]undec-3-en-5-yl]-4-fluorophenyl}-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 2, CHLORIDE ION, N-[3-[(5R)-3-azanyl-5-methyl-9,9-bis(oxidanylidene)-2,9$l^{6}-dithia-4-azaspiro[5.5]undec-3-en-5-yl]-4-fluoranyl-phenyl]-5-(fluoranylmethoxy)pyrazine-2-carboxamide, ... | | Authors: | Fujimoto, K, Matsuoka, E, Asada, N, Tadano, G, Yamamoto, T, Nakahara, K, Fuchino, K, Ito, H, Kanegawa, N, Moechars, D, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structure-Based Design of Selective beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors: Targeting the Flap to Gain Selectivity over BACE2.
J.Med.Chem., 62, 2019
|
|
6JT4
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(4S,6S)-2-amino-4-methyl-6-(trifluoromethyl)-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Anan, K, Iso, Y, Oguma, T, Nakahara, K, Suzuki, S, Yamamoto, T, Matsuoka, E, Ito, H, Sakaguchi, G, Ando, S, Morimoto, K, Kanegawa, N, Kido, Y, Kawachi, T, Fukushima, T, Teisman, A, Urmaliya, V, Dhuyvetter, D, Borghys, H, Austin, N, Bergh, A.V.D, Verboven, P, Bischoff, F, Gijsen, H.J.M, Yamano, Y, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2020-01-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trifluoromethyl Dihydrothiazine-Based beta-Secretase (BACE1) Inhibitors with Robust Central beta-Amyloid Reduction and Minimal Covalent Binding Burden.
Chemmedchem, 14, 2019
|
|
4CWN
 
 | | Human HSP90 alpha N-terminal domain in complex with an Aminotriazoloquinazoline inhibitor | | Descriptor: | 5-(3,5-dimethoxybenzyl)[1,2,4]triazolo[1,5-c]quinazolin-2-amine, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Casale, E, Amboldi, N, Brasca, G, Caronni, D, Colombo, N, Dalvit, C, Felder, E.R, Fogliatto, G, Isacchi, A, Mantegani, S, Polucci, P, Riceputi, L, Sola, F, Visco, C, Zuccotto, F, Casuscelli, F. | | Deposit date: | 2014-04-03 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-Based Hit Discovery and Structure-Based Optimization of Aminotriazoloquinazolines as Novel Hsp90 Inhibitors.
Bioorg.Med.Chem., 22, 2014
|
|
1W8F
 
 | | PSEUDOMONAS AERUGINOSA LECTIN II (PA-IIL)COMPLEXED WITH LACTO-N-NEO- FUCOPENTAOSE V(LNPFV) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Perret, S, Sabin, C, Dumon, C, Budova, M, Gautier, C, Galanina, O, Ilia, S, Bovin, N, Nicaise, M, Desmadril, M, Gilboa-Garber, N, Wimmerova, M, Mitchell, E.P, Imberty, A. | | Deposit date: | 2004-09-21 | | Release date: | 2005-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural Basis for the Interaction between Human Milk Oligosaccharides and the Bacterial Lectin Pa-Iil of Pseudomonas Aeruginosa.
Biochem.J., 389, 2005
|
|
1W8H
 
 | | structure of pseudomonas aeruginosa lectin II (PA-IIL)complexed with lewisA trisaccharide | | Descriptor: | CALCIUM ION, GLYCEROL, PSEUDOMONAS AERUGINOSA LECTIN II, ... | | Authors: | Perret, S, Sabin, C, Dumon, C, Budova, M, Gautier, C, Galanina, O, Ilia, S, Bovin, N, Nicaise, M, Desmadril, M, Gilboa-Garber, N, Wimmerova, M, Mitchell, E.P, Imberty, A. | | Deposit date: | 2004-09-21 | | Release date: | 2005-03-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for the Interaction between Human Milk Oligosaccharides and the Bacterial Lectin Pa-Iil of Pseudomonas Aeruginosa.
Biochem.J., 389, 2005
|
|
1BH6
 
 | | SUBTILISIN DY IN COMPLEX WITH THE SYNTHETIC INHIBITOR N-BENZYLOXYCARBONYL-ALA-PRO-PHE-CHLOROMETHYL KETONE | | Descriptor: | CALCIUM ION, N-BENZYLOXYCARBONYL-ALA-PRO-3-AMINO-4-PHENYL-BUTAN-2-OL, SODIUM ION, ... | | Authors: | Eschenburg, S, Genov, N, Wilson, K.S, Betzel, C. | | Deposit date: | 1998-06-15 | | Release date: | 1998-11-04 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of subtilisin DY, a random mutant of subtilisin Carlsberg.
Eur.J.Biochem., 257, 1998
|
|
6SGY
 
 | |
6ICG
 
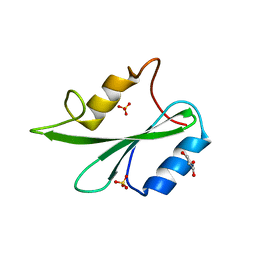 | | Grb2 SH2 domain in phosphopeptide free form | | Descriptor: | GLYCEROL, Growth factor receptor-bound protein 2, SULFATE ION | | Authors: | Hosoe, Y, Numoto, N, Inaba, S, Ogawa, S, Morii, H, Abe, R, Ito, N, Oda, M. | | Deposit date: | 2018-09-06 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural and functional properties of Grb2 SH2 dimer in CD28 binding.
Biophys Physicobio., 16, 2019
|
|
2M64
 
 | | 1H, 13C and 15N Chemical Shift Assignments for Phl p 5a | | Descriptor: | Phlp5 | | Authors: | Goebl, C, Focke, M, Schrank, E, Madl, T, Kosol, S, Madritsch, C, Flicker, S, Valenta, R, Zangger, K, Tjandra, N. | | Deposit date: | 2013-03-21 | | Release date: | 2014-03-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Flexible IgE epitope-containing domains of Phl p 5 cause high allergenic activity.
J. Allergy Clin. Immunol., 140, 2017
|
|
4C7F
 
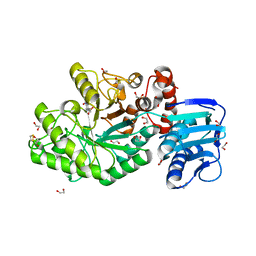 | | Structure and activity of the GH20 beta-N-acetylhexosaminidase from Streptomyces coelicolor A3(2) | | Descriptor: | 1,2-ETHANEDIOL, 6-ACETAMIDO-6-DEOXY-CASTANOSPERMINE, BETA-N-ACETYLHEXOSAMINIDASE | | Authors: | Nguyen Thi, N, Offen, W.A, Davies, G.J, Doucet, N. | | Deposit date: | 2013-09-20 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Activity of the Streptomyces Coelicolor A3(2) Beta-N-Acetylhexosaminidase Provides Further Insight Into Gh20 Family Catalysis and Inhibition.
Biochemistry, 53, 2014
|
|
6SGW
 
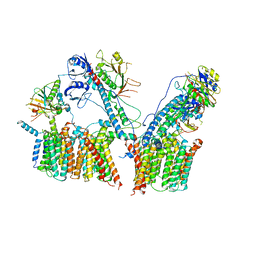 | | Structure of the ESX-3 core complex | | Descriptor: | ESX-3 secretion system ATPase EccB3, ESX-3 secretion system protein EccC3, ESX-3 secretion system protein EccD3, ... | | Authors: | Famelis, N, Rivera-Calzada, A, Llorca, O, Geibel, S. | | Deposit date: | 2019-08-05 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Architecture of the mycobacterial type VII secretion system.
Nature, 576, 2019
|
|
6SX4
 
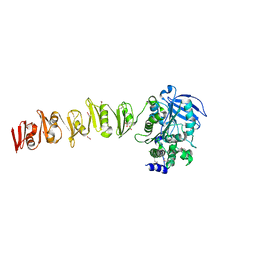 | |
