5O8P
 
 | | The crystal structure of DfoA bound to FAD, the desferrioxamine biosynthetic pathway cadaverine monooxygenase from the fire blight disease pathogen Erwinia amylovora | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-lysine 6-monooxygenase involved in desferrioxamine biosynthesis | | Authors: | Salomone-Stagni, M, Bartho, J.D, Polsinelli, I, Bellini, D, Walsh, M.A, Demitri, N, Benini, S. | | Deposit date: | 2017-06-14 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A complete structural characterization of the desferrioxamine E biosynthetic pathway from the fire blight pathogen Erwinia amylovora.
J.Struct.Biol., 202, 2018
|
|
5OB7
 
 | | X-ray structure of the adduct formed upon reaction of lysozyme with the compound fac-[RuII(CO)3Cl2(N3-IM), IM=imidazole (crystal 2) | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | Pontillo, N, Ferraro, G, Merlino, A. | | Deposit date: | 2017-06-26 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ru-Based CO releasing molecules with azole ligands: interaction with proteins and the CO release mechanism disclosed by X-ray crystallography.
Dalton Trans, 46, 2017
|
|
2Z9Y
 
 | | Crystal structure of CERT START domain in complex with C10-diacylglycerol | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl didecanoate, Lipid-transfer protein CERT | | Authors: | Kudo, N, Kumagai, K, Wakatsuki, S, Nishijima, M, Hanada, K, Kato, R. | | Deposit date: | 2007-09-26 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5OBC
 
 | | X-ray structure of the adduct formed upon reaction of ribonuclease A with the compound fac-[RuII(CO)3Cl2(N3-IM), IM=imidazole | | Descriptor: | PHOSPHATE ION, Ribonuclease pancreatic, pentakis(oxidaniumyl)-(oxidaniumylidynemethyl)ruthenium, ... | | Authors: | Pontillo, N, Ferraro, G, Merlino, A. | | Deposit date: | 2017-06-26 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Ru-Based CO releasing molecules with azole ligands: interaction with proteins and the CO release mechanism disclosed by X-ray crystallography.
Dalton Trans, 46, 2017
|
|
5OIZ
 
 | | Penicillin-Binding Protein 2X (PBP2X) from Streptococcus pneumoniae in complex with oxacillin | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-{[(5-methyl-3-phenyl-1,2-oxazol-4-yl)carbonyl]amino}-2-oxoethyl]-1,3-thiazolidine-4-carb oxylic acid, Penicillin-binding protein 2X | | Authors: | Bernardo-Garcia, N, Hermoso, J.A. | | Deposit date: | 2017-07-20 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Allostery, Recognition of Nascent Peptidoglycan, and Cross-linking of the Cell Wall by the Essential Penicillin-Binding Protein 2x of Streptococcus pneumoniae.
ACS Chem. Biol., 13, 2018
|
|
5O1K
 
 | |
5O95
 
 | |
5OA3
 
 | | Human 40S-eIF2D-re-initiation complex | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Weisser, M, Schaefer, T, Leibundgut, M, Boehringer, D, Aylett, C.H.S, Ban, N. | | Deposit date: | 2017-06-20 | | Release date: | 2017-08-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural and Functional Insights into Human Re-initiation Complexes.
Mol. Cell, 67, 2017
|
|
4M3F
 
 | | Rapid and efficient design of new inhibitors of Mycobacterium tuberculosis transcriptional repressor EthR using fragment growing, merging and linking approaches | | Descriptor: | HTH-type transcriptional regulator EthR, N-(3-methylbutyl)-4-(2-methyl-1,3-thiazol-4-yl)benzenesulfonamide | | Authors: | Villemagne, B, Flipo, M, Blondiaux, N, Crauste, C, Malaquin, S, Leroux, F, Piveteau, C, Villeret, V, Brodin, P, Villoutreix, B, Sperandio, O, Wohlkonig, A, Wintjens, R, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2013-08-06 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand Efficiency Driven Design of New Inhibitors of Mycobacterium tuberculosis Transcriptional Repressor EthR Using Fragment Growing, Merging, and Linking Approaches.
J.Med.Chem., 57, 2014
|
|
5OB9
 
 | | X-ray structure of the adduct formed upon reaction of lysozyme with the compound fac-[RuII(CO)3Cl2(N3-MIM), MIM=methyl-imidazole (crystals grown using ethylene glycol | | Descriptor: | 1,2-ETHANEDIOL, Lysozyme C, NITRATE ION, ... | | Authors: | Pontillo, N, Ferraro, G, Merlino, A. | | Deposit date: | 2017-06-26 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Ru-Based CO releasing molecules with azole ligands: interaction with proteins and the CO release mechanism disclosed by X-ray crystallography.
Dalton Trans, 46, 2017
|
|
5OFL
 
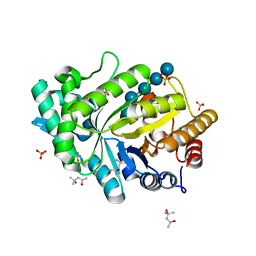 | | Crystal structure of CbXyn10C variant E140Q/E248Q complexed with cellohexaose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Glycoside hydrolase family 48, SULFATE ION, ... | | Authors: | Hakulinen, N, Penttinen, L, Rouvinen, J. | | Deposit date: | 2017-07-11 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.871 Å) | | Cite: | Insights into the roles of non-catalytic residues in the active site of a GH10 xylanase with activity on cellulose.
J. Biol. Chem., 292, 2017
|
|
5OJS
 
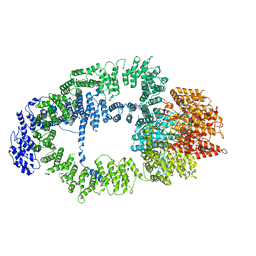 | |
5OFK
 
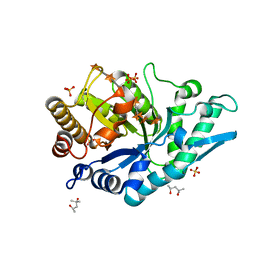 | | Crystal structure of CbXyn10C variant E140Q/E248Q complexed with xyloheptaose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Glycoside hydrolase family 48, PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID), ... | | Authors: | Hakulinen, N, Penttinen, L, Rouvinen, J, Tu, T. | | Deposit date: | 2017-07-11 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Insights into the roles of non-catalytic residues in the active site of a GH10 xylanase with activity on cellulose.
J. Biol. Chem., 292, 2017
|
|
2WZZ
 
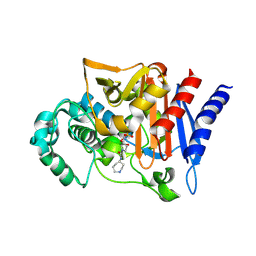 | |
5OFJ
 
 | | Crystal structure of N-terminal domain of bifunctional CbXyn10C | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Glycoside hydrolase family 48 | | Authors: | Hakulinen, N, Penttinen, L, Rouvinen, J. | | Deposit date: | 2017-07-11 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Insights into the roles of non-catalytic residues in the active site of a GH10 xylanase with activity on cellulose.
J. Biol. Chem., 292, 2017
|
|
5OGT
 
 | |
5OJ0
 
 | |
2ZXM
 
 | | A New Class of Vitamin D Receptor Ligands that Induce Structural Rearrangement of the Ligand-binding Pocket | | Descriptor: | (1R,3S,5Z)-5-[(2E)-2-[(1R,3aS,7aR)-1-[(2R,3S)-3-(2-hydroxyethyl)heptan-2-yl]-7a-methyl-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Ikura, T, Ito, N. | | Deposit date: | 2009-01-04 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | A New Class of Vitamin D Analogues that Induce Structural Rearrangement of the Ligand-Binding Pocket of the Receptor
J.Med.Chem., 52, 2009
|
|
3A20
 
 | |
3A3Z
 
 | | Crystal structure of the human VDR ligand binding domain bound to the synthetic agonist compound 2alpha-methyl-AMCR277A(C23S) | | Descriptor: | (1S,2S,3R,5Z,7E,14beta,17alpha)-17-[(2S,4S)-4-(2-hydroxy-2-methylpropyl)-2-methyltetrahydrofuran-2-yl]-2-methyl-9,10-secoandrosta-5,7,10-triene-1,3-diol, SULFATE ION, Vitamin D3 receptor | | Authors: | Sato, Y, Antony, P, Huet, T, Sigueiro, R, Rochel, N, Moras, D, Structural Proteomics in Europe 2 (SPINE-2) | | Deposit date: | 2009-06-25 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure-function relationships and crystal structures of the vitamin D receptor bound 2 alpha-methyl-(20S,23S)- and 2 alpha-methyl-(20S,23R)-epoxymethano-1 alpha,25-dihydroxyvitamin D3
J.Med.Chem., 53, 2010
|
|
4M3D
 
 | | Rapid and efficient design of new inhibitors of Mycobacterium tuberculosis transcriptional repressor EthR using fragment growing, merging and linking approaches | | Descriptor: | 4-{3-[(phenylsulfonyl)amino]prop-1-yn-1-yl}-N-(3,3,3-trifluoropropyl)benzamide, HTH-type transcriptional regulator EthR | | Authors: | Villemagne, B, Flipo, M, Blondiaux, N, Crauste, C, Malaquin, S, Leroux, F, Piveteau, C, Villeret, V, Brodin, P, Villoutreix, B, Sperandio, O, Wohlkonig, A, Wintjens, R, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2013-08-06 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand Efficiency Driven Design of New Inhibitors of Mycobacterium tuberculosis Transcriptional Repressor EthR Using Fragment Growing, Merging, and Linking Approaches.
J.Med.Chem., 57, 2014
|
|
3M1H
 
 | |
2ZIS
 
 | | Crystal Structure of rat protein farnesyltransferase complexed with a bezoruran inhibitor and FPP | | Descriptor: | 3-{2-[(S)-(4-cyanophenyl)(hydroxy)(1-methyl-1H-imidazol-5-yl)methyl]-5-nitro-1-benzofuran-7-yl}benzonitrile, ACETIC ACID, FARNESYL DIPHOSPHATE, ... | | Authors: | Fukami, T.A, Sogabe, S, Nagata, Y, Kondoh, O, Ishii, N. | | Deposit date: | 2008-02-22 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Synthesis and structure-activity relationships of novel benzofuran farnesyltransferase inhibitors
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3W80
 
 | |
3DR1
 
 | | Side-chain fluorine atoms of non-steroidal vitamin D3 analogs stabilize helix 12 of vitamin D receptor | | Descriptor: | (1R,3R)-5-[(2E)-3-{(1S,3R)-2,2,3-trimethyl-3-[6,6,6-trifluoro-5-hydroxy-5-(trifluoromethyl)hex-3-yn-1-yl]cyclopentyl}prop-2-en-1-ylidene]cyclohexane-1,3-diol, MAGNESIUM ION, SRC-1 (LXXLL motif) from Nuclear receptor coactivator 1, ... | | Authors: | Sato, Y, Rochel, N, Moras, D. | | Deposit date: | 2008-07-10 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Superagonistic fluorinated vitamin D3 analogs stabilize helix 12 of the vitamin D receptor.
Chem.Biol., 15, 2008
|
|
