6DEU
 
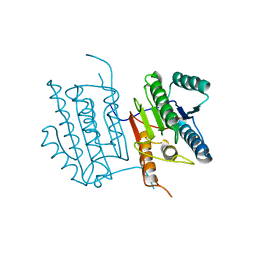 | | Human caspase-6 A109T | | Descriptor: | Caspase-6 | | Authors: | Tubeleviciute-Aydin, A, Beautrait, A, Lynham, J, Sharma, G, Gorelik, A, Deny, L.J, Soya, N, Lukacs, G.L, Nagar, B, Marinier, A, LeBlanc, A.C. | | Deposit date: | 2018-05-13 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Identification of Allosteric Inhibitors against Active Caspase-6.
Sci Rep, 9, 2019
|
|
8WI9
 
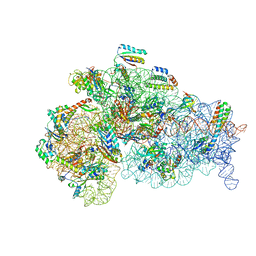 | | Cryo- EM structure of Mycobacterium smegmatis 30S ribosomal subunit (body 2) of 70S ribosome, bS1 and RafH. | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Kumar, N, Sharma, S, Kaushal, P.S. | | Deposit date: | 2023-09-24 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo- EM structure of the mycobacterial 70S ribosome in complex with ribosome hibernation promotion factor RafH.
Nat Commun, 15, 2024
|
|
8WID
 
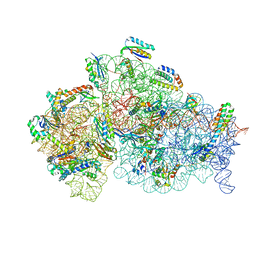 | | Cryo- EM structure of Mycobacterium smegmatis 30S ribosomal subunit (body 2) of 70S ribosome, E- tRNA and RafH. | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Kumar, N, Sharma, S, Kaushal, P.S. | | Deposit date: | 2023-09-24 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo- EM structure of the mycobacterial 70S ribosome in complex with ribosome hibernation promotion factor RafH.
Nat Commun, 15, 2024
|
|
6DS5
 
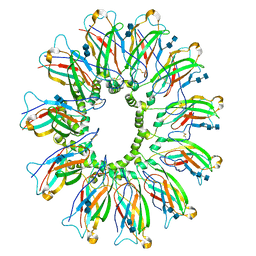 | | Cryo EM structure of human SEIPIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Seipin | | Authors: | Yan, R.H, Qian, H.W, Yan, N, Yang, H.Y. | | Deposit date: | 2018-06-13 | | Release date: | 2018-10-24 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Human SEIPIN Binds Anionic Phospholipids.
Dev. Cell, 47, 2018
|
|
8WHX
 
 | | Cryo- EM structure of Mycobacterium smegmatis 70S ribosome and RafH. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Kumar, N, Sharma, S, Kaushal, P.S. | | Deposit date: | 2023-09-23 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo- EM structure of the mycobacterial 70S ribosome in complex with ribosome hibernation promotion factor RafH.
Nat Commun, 15, 2024
|
|
8WIB
 
 | | Cryo- EM structure of Mycobacterium smegmatis 70S ribosome, E- tRNA and RafH. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Kumar, N, Sharma, S, Kaushal, P.S. | | Deposit date: | 2023-09-24 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo- EM structure of the mycobacterial 70S ribosome in complex with ribosome hibernation promotion factor RafH.
Nat Commun, 15, 2024
|
|
8WHY
 
 | | Cryo- EM structure of Mycobacterium smegmatis 50S ribosomal subunit (body 1) of 70S ribosome and RafH. | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Kumar, N, Sharma, S, Kaushal, P.S. | | Deposit date: | 2023-09-23 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo- EM structure of the mycobacterial 70S ribosome in complex with ribosome hibernation promotion factor RafH.
Nat Commun, 15, 2024
|
|
8WI7
 
 | | Cryo- EM structure of Mycobacterium smegmatis 70S ribosome, bS1 and RafH. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Kumar, N, Sharma, S, Kaushal, P.S. | | Deposit date: | 2023-09-24 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo- EM structure of the mycobacterial 70S ribosome in complex with ribosome hibernation promotion factor RafH.
Nat Commun, 15, 2024
|
|
8WIC
 
 | | Cryo- EM structure of Mycobacterium smegmatis 50S ribosomal subunit (body 1) of 70S ribosome, E- tRNA and RafH. | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Kumar, N, Sharma, S, Kaushal, P.S. | | Deposit date: | 2023-09-24 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo- EM structure of the mycobacterial 70S ribosome in complex with ribosome hibernation promotion factor RafH.
Nat Commun, 15, 2024
|
|
6CAM
 
 | |
8WIF
 
 | | Cryo- EM structure of Mycobacterium smegmatis 30S ribosomal subunit (body 2) of 70S ribosome and RafH. | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Kumar, N, Sharma, S, Kaushal, P.S. | | Deposit date: | 2023-09-24 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo- EM structure of the mycobacterial 70S ribosome in complex with ribosome hibernation promotion factor RafH.
Nat Commun, 15, 2024
|
|
8WI8
 
 | | Cryo- EM structure of Mycobacterium smegmatis 50S ribosomal subunit (body 1) of 70S ribosome, bS1 and RafH. | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Kumar, N, Sharma, S, Kaushal, P.S. | | Deposit date: | 2023-09-24 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo- EM structure of the mycobacterial 70S ribosome in complex with ribosome hibernation promotion factor RafH.
Nat Commun, 15, 2024
|
|
6RV4
 
 | | Crystal structure of the human two pore domain potassium ion channel TASK-1 (K2P3.1) in a closed conformation with a bound inhibitor BAY 2341237 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, POTASSIUM ION, ... | | Authors: | Rodstrom, K.E.J, Pike, A.C.W, Zhang, W, Quigley, A, Speedman, D, Mukhopadhyay, S.M.M, Shrestha, L, Chalk, R, Venkaya, S, Bushell, S.R, Tessitore, A, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-30 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A lower X-gate in TASK channels traps inhibitors within the vestibule.
Nature, 582, 2020
|
|
8XMN
 
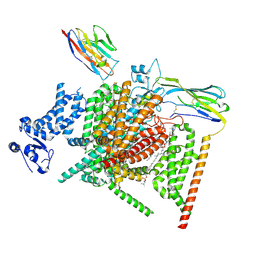 | | Voltage-gated sodium channel Nav1.7 variant M2 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Li, Z, Wu, Q, Huang, G. | | Deposit date: | 2023-12-27 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Dissection of the structure-function relationship of Na v channels.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
2NQZ
 
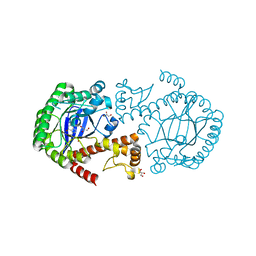 | | Trna-guanine transglycosylase (TGT) mutant in complex with 7-deaza-7-aminomethyl-guanine | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Tidten, N, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2006-11-01 | | Release date: | 2007-11-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor
Plos One, 8, 2013
|
|
7JV2
 
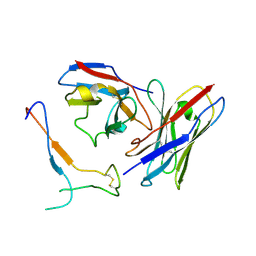 | | SARS-CoV-2 spike in complex with the S2H13 neutralizing antibody Fab fragment (local refinement of the receptor-binding motif and Fab variable domains) | | Descriptor: | S2H13 Fab heavy chain, S2H13 Fab light chain, Spike glycoprotein | | Authors: | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-20 | | Release date: | 2020-10-14 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
6BLK
 
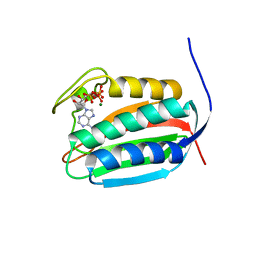 | | Mycobacterial sensor histidine kinase MprB | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Signal transduction histidine-protein kinase/phosphatase mprB | | Authors: | Li, J, Korotkova, N, Korotkov, K.V. | | Deposit date: | 2017-11-10 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Mycobacterial sensor histidine kinase MprB
to be published
|
|
7JW0
 
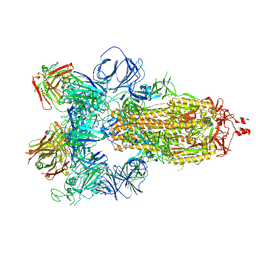 | | SARS-CoV-2 spike in complex with the S304 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S304 Fab heavy chain, ... | | Authors: | Walls, A.C, Park, Y.J, Tortorici, M.A, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-24 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
8XMO
 
 | | Voltage-gated sodium channel Nav1.7 variant M4 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Li, Z, Wu, Q, Huang, G. | | Deposit date: | 2023-12-27 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Dissection of the structure-function relationship of Na v channels.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7JR9
 
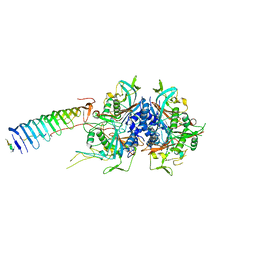 | | Chlamydomonas reinhardtii radial spoke minimal head complex | | Descriptor: | Flagellar radial spoke protein 10, Flagellar radial spoke protein 4, Flagellar radial spoke protein 6, ... | | Authors: | Grossman-Haham, I, Coudray, N, Yu, Z, Wang, F, Bhabha, G, Vale, R.D. | | Deposit date: | 2020-08-11 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structure of the radial spoke head and insights into its role in mechanoregulation of ciliary beating.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6C8G
 
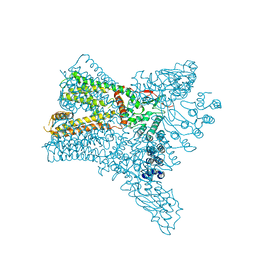 | | Crystal structure of Transient Receptor Potential (TRP) channel TRPV4 in the presence of barium | | Descriptor: | BARIUM ION, Transient receptor potential cation channel, subfamily V, ... | | Authors: | Deng, Z, Paknejad, N, Maksaev, G, Sala-Rabanal, M, Nichols, C.G, Hite, R.K, Yuan, P. | | Deposit date: | 2018-01-24 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (6.31 Å) | | Cite: | Cryo-EM and X-ray structures of TRPV4 reveal insight into ion permeation and gating mechanisms.
Nat. Struct. Mol. Biol., 25, 2018
|
|
8UF7
 
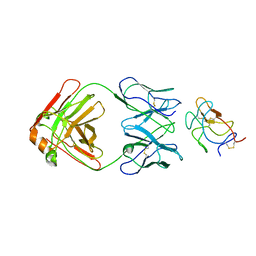 | | Cryo-EM structure of POmAb, a Type-I anti-prothrombin antiphospholipid antibody, bound to kringle-1 of human prothrombin | | Descriptor: | POmAb Heavy Chain, POmAb Light Chain, Prothrombin | | Authors: | Kumar, S, Summers, B, Basore, K, Pozzi, N. | | Deposit date: | 2023-10-03 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure and functional basis of prothrombin recognition by a type I antiprothrombin antiphospholipid antibody.
Blood, 143, 2024
|
|
1X65
 
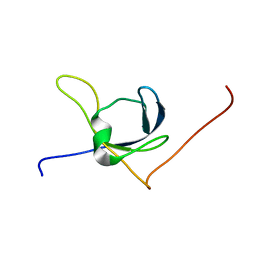 | | Solution structure of the third cold-shock domain of the human KIAA0885 protein (UNR PROTEIN) | | Descriptor: | UNR protein | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-17 | | Release date: | 2005-11-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structures of the five constituent cold-shock domains (CSD) of the human UNR (upstream of N-ras) protein.
J.Struct.Funct.Genom., 11, 2010
|
|
8XMM
 
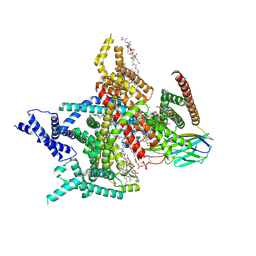 | | Voltage-gated sodium channel Nav1.7 variant M9 | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Yan, N, Li, Z, Wu, Q, Huang, G. | | Deposit date: | 2023-12-27 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Dissection of the structure-function relationship of Na v channels.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6BZW
 
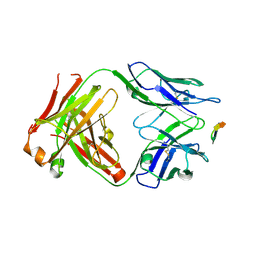 | | Structure of the Hepatitis C virus envelope glycoprotein E2 antigenic region 412-423 bound to the GL precursor of the broadly neutralizing antibody AP33 | | Descriptor: | AP33 GL Heavy Chain, AP33 GL Light Chain, E2 AS412 peptide | | Authors: | Tzarum, N, Aleman, F, Wilson, I.A, Law, M. | | Deposit date: | 2017-12-26 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Immunogenetic and structural analysis of a class of HCV broadly neutralizing antibodies and their precursors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
