4V3P
 
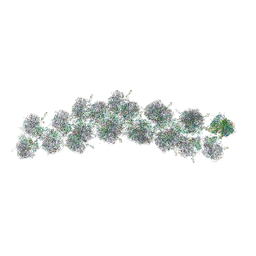 | | The molecular structure of the left-handed supra-molecular helix of eukaryotic polyribosomes | | Descriptor: | 18S ribosomal RNA, 26S ribosomal RNA, 40S WHEAT GERM RIBOSOME protein 4, ... | | Authors: | Myasnikov, A.G, Afonina, Z.A, Menetret, J.F, Shirokov, V.A, Spirin, A.S, Klaholz, B.P. | | Deposit date: | 2014-10-20 | | Release date: | 2015-04-22 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (34 Å) | | Cite: | The molecular structure of the left-handed supra-molecular helix of eukaryotic polyribosomes.
Nat Commun, 5, 2014
|
|
5LKS
 
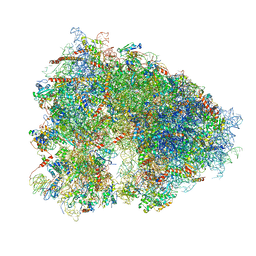 | | Structure-function insights reveal the human ribosome as a cancer target for antibiotics | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Myasnikov, A.G, Natchiar, S.K, Nebout, M, Hazemann, I, Imbert, V, Khatter, H, Peyron, J.-F, Klaholz, B.P. | | Deposit date: | 2016-07-23 | | Release date: | 2017-04-26 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure-function insights reveal the human ribosome as a cancer target for antibiotics.
Nat Commun, 7, 2016
|
|
7LHT
 
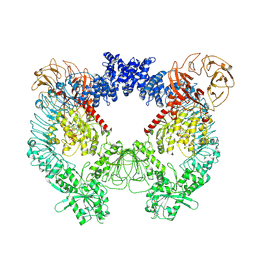 | | Structure of the LRRK2 dimer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Myasnikov, A, Zhu, H, Hixson, P, Xie, B, Yu, K, Pitre, A, Peng, J, Sun, J. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural analysis of the full-length human LRRK2.
Cell, 184, 2021
|
|
7LHW
 
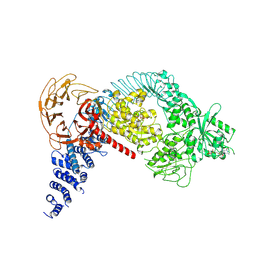 | | Structure of the LRRK2 monomer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Myasnikov, A, Zhu, H, Hixson, P, Xie, B, Yu, K, Pitre, A, Peng, J, Sun, J. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural analysis of the full-length human LRRK2.
Cell, 184, 2021
|
|
7LI3
 
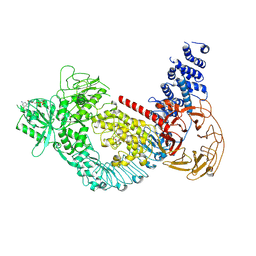 | | Structure of the LRRK2 G2019S mutant | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Myasnikov, A, Zhu, H, Hixson, P, Xie, B, Yu, K, Pitre, A, Peng, J, Sun, J. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural analysis of the full-length human LRRK2.
Cell, 184, 2021
|
|
7LI4
 
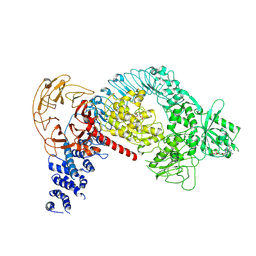 | | Structure of LRRK2 after symmetry expansion | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Myasnikov, A, Zhu, H, Hixson, P, Xie, B, Yu, K, Pitre, A, Peng, J, Sun, J. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural analysis of the full-length human LRRK2.
Cell, 184, 2021
|
|
7LVK
 
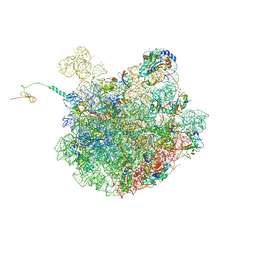 | | Cfr-modified 50S subunit from Escherichia coli | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Stojkovic, V, Myasnikov, A.G, Frost, A, Fujimori, D.G. | | Deposit date: | 2021-02-25 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Investigating antibiotic resistance of a ribosomal-RNA methylating enzyme through directed evolution
To Be Published
|
|
8FWK
 
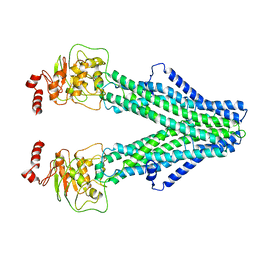 | |
3J4J
 
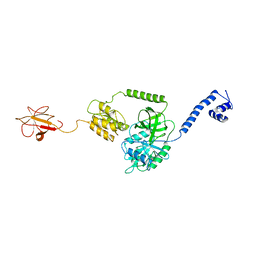 | | Model of full-length T. thermophilus Translation Initiation Factor 2 refined against its cryo-EM density from a 30S Initiation Complex map | | Descriptor: | Translation initiation factor IF-2 | | Authors: | Simonetti, A, Marzi, S, Billas, I.M.L, Tsai, A, Fabbretti, A, Myasnikov, A, Roblin, P, Vaiana, A.C, Hazemann, I, Eiler, D, Steitz, T.A, Puglisi, J.D, Gualerzi, C.O, Klaholz, B.P. | | Deposit date: | 2013-08-26 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Involvement of protein IF2 N domain in ribosomal subunit joining revealed from architecture and function of the full-length initiation factor.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7QO9
 
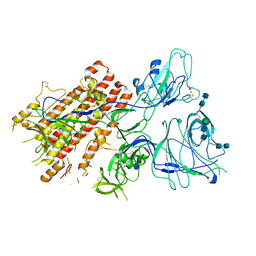 | | SARS-CoV-2 S Omicron Spike B.1.1.529 - RBD and NTD (Local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,SARS-CoV-2 S Omicron Spike B.1.1.529, ... | | Authors: | Ni, D, Lau, K, Turelli, P, Beckert, B, Nazarov, S, Pojer, F, Myasnikov, A, Stahlberg, H, Trono, D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-26 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Structural analysis of the Spike of the Omicron SARS-COV-2 variant by cryo-EM and implications for immune evasion
Biorxiv, 2021
|
|
6QZP
 
 | | High-resolution cryo-EM structure of the human 80S ribosome | | Descriptor: | (3beta)-O~3~-[(2R)-2,6-dihydroxy-2-(2-methoxy-2-oxoethyl)-6-methylheptanoyl]cephalotaxine, 18S rRNA (1740-MER), 28S rRNA (3773-MER), ... | | Authors: | Natchiar, S.K, Myasnikov, A.G, Kratzat, H, Hazemann, I, Klaholz, B.P. | | Deposit date: | 2019-03-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Visualization of chemical modifications in the human 80S ribosome structure.
Nature, 551, 2017
|
|
2VAZ
 
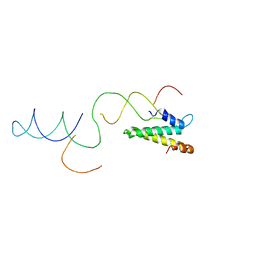 | | Model of the S15-mRNA complex fitted into the cryo-EM map of the 70S entrapment complex. | | Descriptor: | 30S RIBOSOMAL PROTEIN S15, RPSO MRNA OPERATOR | | Authors: | Marzi, S, Myasnikov, A.G, Serganov, A, Ehresmann, C, Romby, P, Yusupov, M, Klaholz, B.P. | | Deposit date: | 2007-09-05 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structured Mrnas Regulate Translation Initiation by Binding to the Platform of the Ribosome.
Cell(Cambridge,Mass.), 130, 2007
|
|
7QO7
 
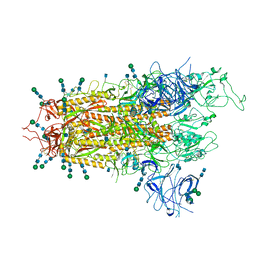 | | SARS-CoV-2 S Omicron Spike B.1.1.529 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ni, D, Lau, K, Turelli, P, Beckert, B, Nazarov, S, Pojer, F, Myasnikov, A, Stahlberg, H, Trono, D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural analysis of the Spike of the Omicron SARS-COV-2 variant by cryo-EM and implications for immune evasion
Biorxiv, 2021
|
|
1ML5
 
 | | Structure of the E. coli ribosomal termination complex with release factor 2 | | Descriptor: | 30S 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Klaholz, B.P, Pape, T, Zavialov, A.V, Myasnikov, A.G, Orlova, E.V, Vestergaard, B, Ehrenberg, M, van Heel, M. | | Deposit date: | 2002-08-30 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Structure of the Escherichia coli ribosomal termination complex with release factor 2
Nature, 421, 2003
|
|
8RQB
 
 | | Cryo-EM structure of mouse heavy-chain apoferritin | | Descriptor: | FE (III) ION, Ferritin heavy chain, N-terminally processed, ... | | Authors: | Nazarov, S, Myasnikov, A, Stahlberg, H. | | Deposit date: | 2024-01-17 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (1.09 Å) | | Cite: | Low-dose cryo-electron ptychography of proteins at sub-nanometer resolution.
Nat Commun, 15, 2024
|
|
6BQR
 
 | | Human TRPM4 ion channel in lipid nanodiscs in a calcium-free state | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Transient receptor potential cation channel subfamily M member 4 | | Authors: | Autzen, H.E, Myasnikov, A.G, Campbell, M.G, Asarnow, D, Julius, D, Cheng, Y. | | Deposit date: | 2017-11-28 | | Release date: | 2017-12-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human TRPM4 ion channel in a lipid nanodisc.
Science, 359, 2018
|
|
6BQV
 
 | | Human TRPM4 ion channel in lipid nanodiscs in a calcium-bound state | | Descriptor: | CALCIUM ION, CHOLESTEROL HEMISUCCINATE, Transient receptor potential cation channel subfamily M member 4 | | Authors: | Autzen, H.E, Myasnikov, A.G, Campbell, M.G, Asarnow, D, Julius, D, Cheng, Y. | | Deposit date: | 2017-11-28 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the human TRPM4 ion channel in a lipid nanodisc.
Science, 359, 2018
|
|
6PJ6
 
 | | High resolution cryo-EM structure of E.coli 50S | | Descriptor: | 23S rRNA, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | Authors: | Stojkovic, V, Myasnikov, A, Frost, A, Fujimori, D.G. | | Deposit date: | 2019-06-27 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Assessment of the nucleotide modifications in the high-resolution cryo-electron microscopy structure of the Escherichia coli 50S subunit.
Nucleic Acids Res., 48, 2020
|
|
8EAL
 
 | | SsoMCM hexamer bound to Mg/ADP-BeFx and DNA. Class 1. Merged particles from datasets with 3 different DNA entities | | Descriptor: | DNA, MAGNESIUM ION, Minichromosome maintenance protein MCM, ... | | Authors: | Meagher, M, Myasnikov, A, Enemark, E.J. | | Deposit date: | 2022-08-29 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | Two Distinct Modes of DNA Binding by an MCM Helicase Enable DNA Translocation.
Int J Mol Sci, 23, 2022
|
|
8EAI
 
 | | SsoMCM hexamer bound to Mg/ADP-BeFx and 16-mer oligo-dT. Class 2 | | Descriptor: | 16-mer oligo-dT, MAGNESIUM ION, Minichromosome maintenance protein MCM, ... | | Authors: | Meagher, M, Myasnikov, A, Enemark, E.J. | | Deposit date: | 2022-08-29 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Two Distinct Modes of DNA Binding by an MCM Helicase Enable DNA Translocation.
Int J Mol Sci, 23, 2022
|
|
8EAK
 
 | | SsoMCM hexamer bound to Mg/ADP-BeFx and 46-mer DNA strand. Class 2 | | Descriptor: | 46-mer DNA, MAGNESIUM ION, Minichromosome maintenance protein MCM, ... | | Authors: | Meagher, M, Myasnikov, A, Enemark, E.J. | | Deposit date: | 2022-08-29 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Two Distinct Modes of DNA Binding by an MCM Helicase Enable DNA Translocation.
Int J Mol Sci, 23, 2022
|
|
8EAM
 
 | | SsoMCM hexamer bound to Mg/ADP-BeFx and DNA. Class 2. Merged particles from datasets with 3 different DNA entities | | Descriptor: | DNA, MAGNESIUM ION, Minichromosome maintenance protein MCM, ... | | Authors: | Meagher, M, Myasnikov, A, Enemark, E.J. | | Deposit date: | 2022-08-29 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Two Distinct Modes of DNA Binding by an MCM Helicase Enable DNA Translocation.
Int J Mol Sci, 23, 2022
|
|
8EAJ
 
 | | SsoMCM hexamer bound to Mg/ADP-BeFx and 46-mer DNA strand. Class 1 | | Descriptor: | 46-mer DNA strand, MAGNESIUM ION, Minichromosome maintenance protein MCM, ... | | Authors: | Meagher, M, Myasnikov, A, Enemark, E.J. | | Deposit date: | 2022-08-29 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Two Distinct Modes of DNA Binding by an MCM Helicase Enable DNA Translocation.
Int J Mol Sci, 23, 2022
|
|
8EAH
 
 | | SsoMCM hexamer bound to Mg/ADP-BeFx and 16-mer oligo-dT. Class 1 | | Descriptor: | 16-mer oligo-dT, MAGNESIUM ION, Minichromosome maintenance protein MCM, ... | | Authors: | Meagher, M, Myasnikov, A, Enemark, E.J. | | Deposit date: | 2022-08-29 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Two Distinct Modes of DNA Binding by an MCM Helicase Enable DNA Translocation.
Int J Mol Sci, 23, 2022
|
|
8EAF
 
 | | SsoMCM hexamer bound to Mg/ADP-BeFx and 12-mer oligo-dT. Class 1 | | Descriptor: | 12-mer oligo dT, MAGNESIUM ION, Minichromosome maintenance protein MCM, ... | | Authors: | Meagher, M, Myasnikov, A, Enemark, E.J. | | Deposit date: | 2022-08-29 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Two Distinct Modes of DNA Binding by an MCM Helicase Enable DNA Translocation.
Int J Mol Sci, 23, 2022
|
|
