1J0M
 
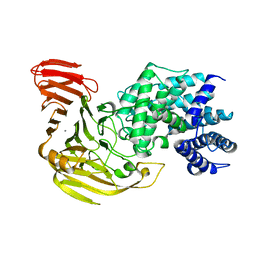 | | Crystal Structure of Bacillus sp. GL1 Xanthan Lyase that Acts on Side Chains of Xanthan | | Descriptor: | CALCIUM ION, XANTHAN LYASE | | Authors: | Hashimoto, W, Nankai, H, Mikami, B, Murata, K. | | Deposit date: | 2002-11-19 | | Release date: | 2003-04-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Bacillus sp. GL1 Xanthan Lyase, Which Acts on the Side Chains of Xanthan.
J.Biol.Chem., 278, 2003
|
|
4TQU
 
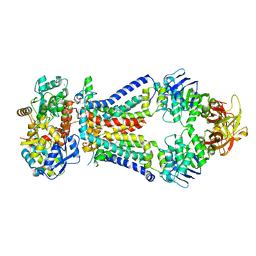 | | Crystal structure of a bacterial ABC transporter involved in the import of the acidic polysaccharide alginate | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, AlgM1, AlgM2, ... | | Authors: | Maruyama, Y, Itoh, T, Kaneko, A, Nishitani, Y, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2014-06-12 | | Release date: | 2015-07-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.204 Å) | | Cite: | Structure of a Bacterial ABC Transporter Involved in the Import of an Acidic Polysaccharide Alginate
Structure, 23, 2015
|
|
4TKL
 
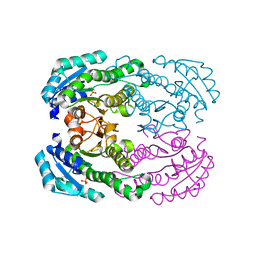 | | Crystal structure of NADH-dependent reductase A1-R' responsible for alginate metabolism | | Descriptor: | NADH-dependent reductase for 4-deoxy-L-erythro-5-hexoseulose uronate, PHOSPHATE ION | | Authors: | Takase, R, Mikami, B, Kawai, S, Murata, K, Hashimoto, W. | | Deposit date: | 2014-05-27 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based Conversion of the Coenzyme Requirement of a Short-chain Dehydrogenase/Reductase Involved in Bacterial Alginate Metabolism.
J.Biol.Chem., 289, 2014
|
|
4TKM
 
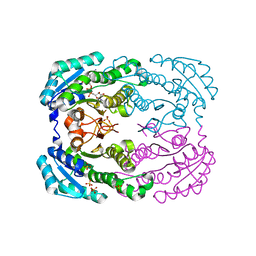 | | Crystal structure of NADH-dependent reductase A1-R' complexed with NAD | | Descriptor: | NADH-dependent reductase for 4-deoxy-L-erythro-5-hexoseulose uronate, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Takase, R, Mikami, B, Kawai, S, Murata, K, Hashimoto, W. | | Deposit date: | 2014-05-27 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structure-based Conversion of the Coenzyme Requirement of a Short-chain Dehydrogenase/Reductase Involved in Bacterial Alginate Metabolism.
J.Biol.Chem., 289, 2014
|
|
4U8F
 
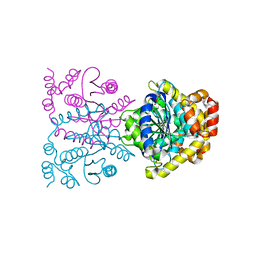 | | Crystal structure of 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase complexed with a tartrate | | Descriptor: | L(+)-TARTARIC ACID, Putative uncharacterized protein gbs1892 | | Authors: | Maruyama, Y, Oiki, S, Takase, R, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Metabolic Fate of Unsaturated Glucuronic/Iduronic Acids from Glycosaminoglycans: MOLECULAR IDENTIFICATION AND STRUCTURE DETERMINATION OF STREPTOCOCCAL ISOMERASE AND DEHYDROGENASE.
J.Biol.Chem., 290, 2015
|
|
4U8G
 
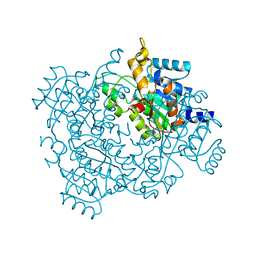 | | Crystal structure of 2-keto-3-deoxy-D-gluconate dehydrogenase from Streptococcus agalactiae | | Descriptor: | Putative uncharacterized protein gbs1891 | | Authors: | Maruyama, Y, Oiki, S, Takase, R, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Metabolic Fate of Unsaturated Glucuronic/Iduronic Acids from Glycosaminoglycans: MOLECULAR IDENTIFICATION AND STRUCTURE DETERMINATION OF STREPTOCOCCAL ISOMERASE AND DEHYDROGENASE.
J.Biol.Chem., 290, 2015
|
|
4U8E
 
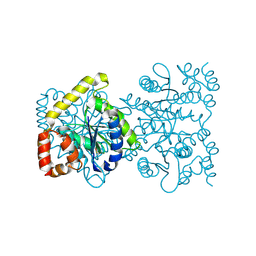 | | Crystal structure of 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase from Streptococcus agalactiae | | Descriptor: | Putative uncharacterized protein gbs1892 | | Authors: | Maruyama, y, Oiki, S, Takase, R, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2014-08-03 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metabolic Fate of Unsaturated Glucuronic/Iduronic Acids from Glycosaminoglycans: MOLECULAR IDENTIFICATION AND STRUCTURE DETERMINATION OF STREPTOCOCCAL ISOMERASE AND DEHYDROGENASE
J.Biol.Chem., 290, 2015
|
|
2AT9
 
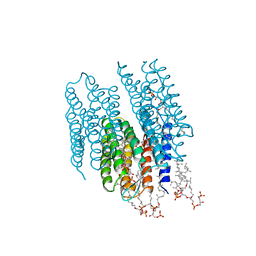 | | STRUCTURE OF BACTERIORHODOPSIN AT 3.0 ANGSTROM BY ELECTRON CRYSTALLOGRAPHY | | Descriptor: | 3-[[3-METHYLPHOSPHONO-GLYCEROLYL]PHOSPHONYL]-[1,2-DI[2,6,10,14-TETRAMETHYL-HEXADECAN-16-YL]GLYCEROL, BACTERIORHODOPSIN, RETINAL | | Authors: | Mitsuoka, K, Hirai, T, Murata, K, Miyazawa, A, Kidera, A, Kimura, Y, Fujiyoshi, Y. | | Deposit date: | 1998-12-17 | | Release date: | 1999-04-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (3 Å) | | Cite: | The structure of bacteriorhodopsin at 3.0 A resolution based on electron crystallography: implication of the charge distribution.
J.Mol.Biol., 286, 1999
|
|
5GX7
 
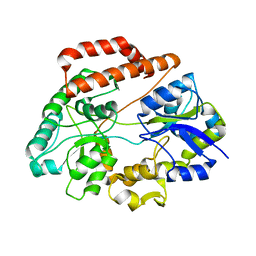 | | Crystal structure of solute-binding protein complexed with unsaturated chondroitin disaccharide with a sulfate group at C-6 position of GalNAc | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-6-O-sulfo-beta-D-galactopyranose, CALCIUM ION, Extracellular solute-binding protein family 1 | | Authors: | Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2016-09-15 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | A bacterial ABC transporter enables import of mammalian host glycosaminoglycans
Sci Rep, 7, 2017
|
|
5GUB
 
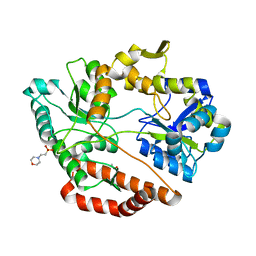 | | Crystal structure of solute-binding protein complexed with sulfate group-free unsaturated chondroitin disaccharide | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, ... | | Authors: | Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2016-08-26 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A bacterial ABC transporter enables import of mammalian host glycosaminoglycans
Sci Rep, 7, 2017
|
|
5GX6
 
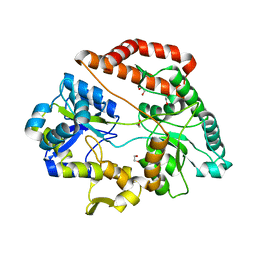 | | Crystal structure of solute-binding protein complexed with unsaturated chondroitin disaccharide with a sulfate group at C-4 position of GalNAc | | Descriptor: | 1,2-ETHANEDIOL, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, CALCIUM ION, ... | | Authors: | Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2016-09-15 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A bacterial ABC transporter enables import of mammalian host glycosaminoglycans
Sci Rep, 7, 2017
|
|
4Z9Y
 
 | | Crystal structure of 2-keto-3-deoxy-D-gluconate dehydrogenase from Pectobacterium carotovorum | | Descriptor: | 2-deoxy-D-gluconate 3-dehydrogenase, SULFATE ION | | Authors: | Takase, R, Maruyama, Y, Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2015-04-13 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural determinants in bacterial 2-keto-3-deoxy-D-gluconate dehydrogenase KduD for dual-coenzyme specificity
Proteins, 84, 2016
|
|
1Y3I
 
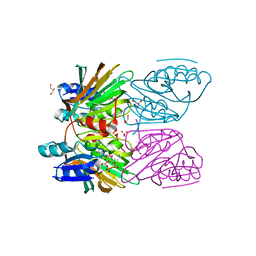 | | Crystal Structure of Mycobacterium tuberculosis NAD kinase-NAD complex | | Descriptor: | GLYCEROL, Inorganic polyphosphate/ATP-NAD kinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Mori, S, Yamasaki, M, Maruyama, Y, Momma, K, Kawai, S, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2004-11-25 | | Release date: | 2005-01-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | NAD-binding mode and the significance of intersubunit contact revealed by the crystal structure of Mycobacterium tuberculosis NAD kinase-NAD complex
Biochem.Biophys.Res.Commun., 327, 2005
|
|
1Y3Q
 
 | | Structure of AlgQ1, alginate-binding protein | | Descriptor: | AlgQ1, CALCIUM ION | | Authors: | Momma, K, Mishima, Y, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2004-11-26 | | Release date: | 2005-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Direct Evidence for Sphingomonas sp. A1 Periplasmic Proteins as Macromolecule-Binding Proteins Associated with the ABC Transporter: Molecular Insights into Alginate Transport in the Periplasm(,)
Biochemistry, 44, 2005
|
|
1Y3P
 
 | | Structure of AlgQ1, alginate-binding protein, complexed with an alginate tetrasaccharide | | Descriptor: | AlgQ1, CALCIUM ION, beta-D-mannopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid | | Authors: | Momma, K, Mishima, Y, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2004-11-26 | | Release date: | 2005-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Direct Evidence for Sphingomonas sp. A1 Periplasmic Proteins as Macromolecule-Binding Proteins Associated with the ABC Transporter: Molecular Insights into Alginate Transport in the Periplasm(,)
Biochemistry, 44, 2005
|
|
1Y3N
 
 | | Structure of AlgQ1, alginate-binding protein, complexed with an alginate disaccharide | | Descriptor: | AlgQ1, CALCIUM ION, beta-D-mannopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid | | Authors: | Momma, K, Mishima, Y, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2004-11-26 | | Release date: | 2005-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Direct Evidence for Sphingomonas sp. A1 Periplasmic Proteins as Macromolecule-Binding Proteins Associated with the ABC Transporter: Molecular Insights into Alginate Transport in the Periplasm(,)
Biochemistry, 44, 2005
|
|
1Y3H
 
 | | Crystal Structure of Inorganic Polyphosphate/ATP-NAD kinase from Mycobacterium tuberculosis | | Descriptor: | Inorganic polyphosphate/ATP-NAD kinase | | Authors: | Mori, S, Yamasaki, M, Maruyama, Y, Momma, K, kawai, S, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2004-11-24 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | NAD-binding mode and the significance of intersubunit contact revealed by the crystal structure of Mycobacterium tuberculosis NAD kinase-NAD complex
BIOCHEM.BIOPHYS.RES.COMMUN., 327, 2005
|
|
5Y4C
 
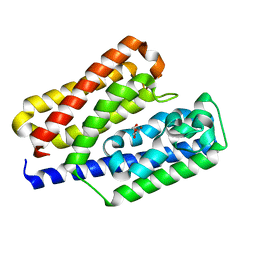 | | Crystal structure of EfeO-like protein Algp7 in complex with a metal ion | | Descriptor: | Alginate-binding protein, COPPER (II) ION, GLYCEROL | | Authors: | Temtrirath, K, Maruyama, Y, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2017-08-03 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Binding mode of metal ions to the bacterial iron import protein EfeO
Biochem. Biophys. Res. Commun., 493, 2017
|
|
4ZA2
 
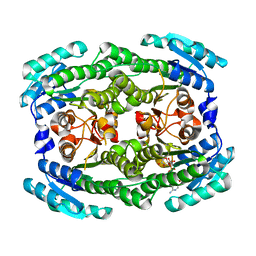 | | Crystal structure of Pectobacterium carotovorum 2-keto-3-deoxy-D-gluconate dehydrogenase complexed with NAD+ | | Descriptor: | 2-deoxy-D-gluconate 3-dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Takase, R, Maruyama, Y, Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2015-04-13 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural determinants in bacterial 2-keto-3-deoxy-D-gluconate dehydrogenase KduD for dual-coenzyme specificity
Proteins, 84, 2016
|
|
4Z9X
 
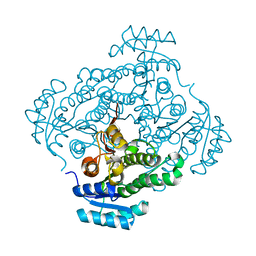 | | Crystal structure of 2-keto-3-deoxy-D-gluconate dehydrogenase from Streptococcus pyogenes | | Descriptor: | Gluconate 5-dehydrogenase | | Authors: | Maruyama, Y, Takase, R, Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2015-04-13 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural determinants in bacterial 2-keto-3-deoxy-D-gluconate dehydrogenase KduD for dual-coenzyme specificity
Proteins, 84, 2016
|
|
6A6U
 
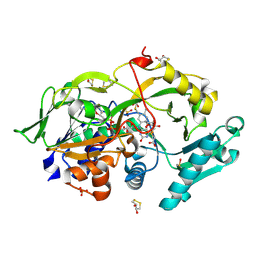 | | Crystal structure of the modified fructosyl peptide oxidase from Aspergillus nidulans with R61G mutation, in complex with FSA | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, 1-S-(carboxymethyl)-1-thio-beta-D-fructopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Ogawa, N, Maruyama, Y, Itoh, T, Hashimoto, W, Murata, K. | | Deposit date: | 2018-06-29 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.945 Å) | | Cite: | Creation of haemoglobin A1c direct oxidase from fructosyl peptide oxidase by combined structure-based site specific mutagenesis and random mutagenesis.
Sci Rep, 9, 2019
|
|
6A6T
 
 | | Crystal structure of the modified fructosyl peptide oxidase from Aspergillus nidulans with R61G mutation | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl amine: oxygen oxidoreductase, ... | | Authors: | Ogawa, N, Maruyama, Y, Itoh, T, Hashimoto, W, Murata, K. | | Deposit date: | 2018-06-29 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Creation of haemoglobin A1c direct oxidase from fructosyl peptide oxidase by combined structure-based site specific mutagenesis and random mutagenesis.
Sci Rep, 9, 2019
|
|
7WMP
 
 | | Tail structure of Helicobacter pylori bacteriophage KHP30 | | Descriptor: | Adaptor protein gp12, Nozzle protein gp25, Portal protein | | Authors: | Kamiya, R, Uchiyama, J, Matsuzaki, S, Murata, K, Iwasaki, K, Miyazaki, N. | | Deposit date: | 2022-01-15 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of Helicobacter pylori bacteriophage KHP30
To Be Published
|
|
5Y7O
 
 | | Crystal structure of folding sensor region of UGGT from Thermomyces dupontii | | Descriptor: | UGGT | | Authors: | Satoh, T, Song, C, Zhu, T, Toshimori, T, Murata, K, Hayashi, Y, Kamikubo, H, Uchihashi, T, Kato, K. | | Deposit date: | 2017-08-17 | | Release date: | 2017-09-27 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Visualisation of a flexible modular structure of the ER folding-sensor enzyme UGGT.
Sci Rep, 7, 2017
|
|
6A6V
 
 | | Crystal structure of the modified fructosyl peptide oxidase from Aspergillus nidulans with 7 additional mutations, in complex with FSA | | Descriptor: | 1-S-(carboxymethyl)-1-thio-beta-D-fructopyranose, FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl amine: oxygen oxidoreductase | | Authors: | Ogawa, N, Maruyama, Y, Itoh, T, Hashimoto, W, Murata, K. | | Deposit date: | 2018-06-29 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Creation of haemoglobin A1c direct oxidase from fructosyl peptide oxidase by combined structure-based site specific mutagenesis and random mutagenesis.
Sci Rep, 9, 2019
|
|
