4JK2
 
 | |
2Z5I
 
 | | Crystal structure of the head-to-tail junction of tropomyosin | | Descriptor: | General control protein GCN4 and Tropomyosin alpha-1 chain, MAGNESIUM ION, Tropomyosin alpha-1 chain and General control protein GCN4 | | Authors: | Murakami, K, Nozawa, K, Tomii, K, Kudou, N, Igarashi, N, Shirakihara, Y, Wakatsuki, S, Stewart, M, Yasunaga, T, Wakabayashi, T. | | Deposit date: | 2007-07-12 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for tropomyosin overlap in thin (actin) filaments and the generation of a molecular swivel by troponin-T
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
4KMU
 
 | |
2Z5H
 
 | | Crystal structure of the head-to-tail junction of tropomyosin complexed with a fragment of TnT | | Descriptor: | General control protein GCN4 and Tropomyosin alpha-1 chain, Tropomyosin alpha-1 chain and General control protein GCN4, Troponin T, ... | | Authors: | Murakami, K, Nozawa, K, Tomii, K, Kudou, N, Igarashi, N, Shirakihara, Y, Wakatsuki, S, Stewart, M, Yasunaga, T, Wakabayashi, T. | | Deposit date: | 2007-07-12 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural basis for tropomyosin overlap in thin (actin) filaments and the generation of a molecular swivel by troponin-T
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
4KN4
 
 | |
4KN7
 
 | |
3P8B
 
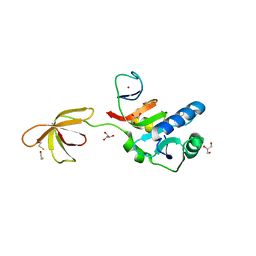 | |
3Q0A
 
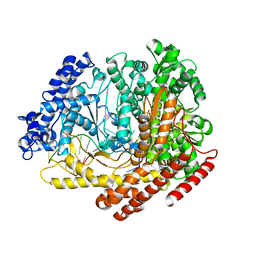 | |
8EMB
 
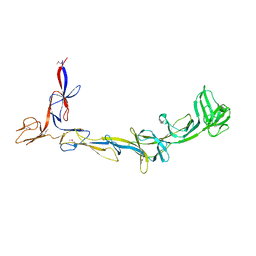 | | X-ray crystal structure of Thermosynechococcus elongatus Si3 domain of RNA polymerase RpoC2 subunit | | Descriptor: | DNA-directed RNA polymerase subunit beta' | | Authors: | Murakami, K.S, Imashimizu, M, Qayyum, M.Z, Vishwakarma, R.K, Yuzenkova, Y. | | Deposit date: | 2022-09-27 | | Release date: | 2022-11-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structure and function of the Si3 insertion integrated into the trigger loop/helix of cyanobacterial RNA polymerase.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
3A5M
 
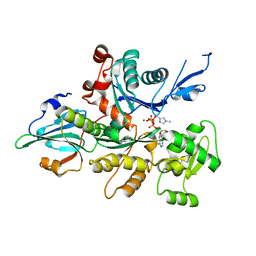 | | Crystal Structure of a Dictyostelium P109I Mg2+-Actin in Complex with Human Gelsolin Segment 1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Gelsolin, ... | | Authors: | Murakami, K, Yasunaga, T, Noguchi, T.Q, Uyeda, T.Q, Wakabayashi, T. | | Deposit date: | 2009-08-09 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for actin assembly, activation of ATP hydrolysis, and delayed phosphate release
Cell(Cambridge,Mass.), 143, 2010
|
|
3A5L
 
 | | Crystal Structure of a Dictyostelium P109A Mg2+-Actin in Complex with Human Gelsolin Segment 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Gelsolin, ... | | Authors: | Murakami, K, Yasunaga, T, Noguchi, T.Q, Uyeda, T.Q, Wakabayashi, T. | | Deposit date: | 2009-08-09 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for actin assembly, activation of ATP hydrolysis, and delayed phosphate release
Cell(Cambridge,Mass.), 143, 2010
|
|
3A5O
 
 | | Crystal Structure of a Dictyostelium P109I Ca2+-Actin in Complex with Human Gelsolin Segment 1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Gelsolin, ... | | Authors: | Murakami, K, Yasunaga, T, Noguchi, T.Q, Uyeda, T.Q, Wakabayashi, T. | | Deposit date: | 2009-08-09 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for actin assembly, activation of ATP hydrolysis, and delayed phosphate release
Cell(Cambridge,Mass.), 143, 2010
|
|
3A5N
 
 | | Crystal Structure of a Dictyostelium P109A Ca2+-Actin in Complex with Human Gelsolin Segment 1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Gelsolin, ... | | Authors: | Murakami, K, Yasunaga, T, Noguchi, T.Q, Uyeda, T.Q, Wakabayashi, T. | | Deposit date: | 2009-08-09 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural basis for actin assembly, activation of ATP hydrolysis, and delayed phosphate release
Cell(Cambridge,Mass.), 143, 2010
|
|
4FF3
 
 | | N4 mini-vRNAP transcription initiation complex, 3 min after soaking GTP, ATP and Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Bacteriophage N4 P2 promoter, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Murakami, K.S, Basu, R.S. | | Deposit date: | 2012-05-30 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Watching the Bacteriophage N4 RNA Polymerase Transcription by Time-dependent Soak-trigger-freeze X-ray Crystallography.
J.Biol.Chem., 288, 2013
|
|
4FF2
 
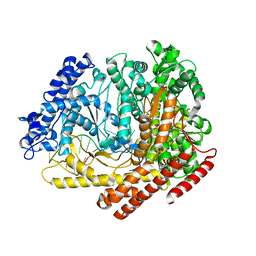 | | N4 mini-vRNAP transcription initiation complex, 2 min after soaking GTP, ATP and Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Bacteriophage N4 P2 promoter, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Murakami, K.S, Basu, R.S. | | Deposit date: | 2012-05-30 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Watching the Bacteriophage N4 RNA Polymerase Transcription by Time-dependent Soak-trigger-freeze X-ray Crystallography.
J.Biol.Chem., 288, 2013
|
|
4FF4
 
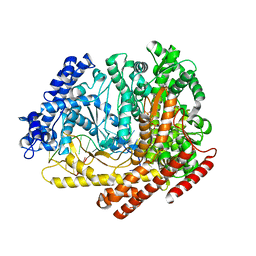 | |
4FF1
 
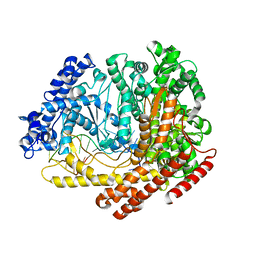 | |
1VDJ
 
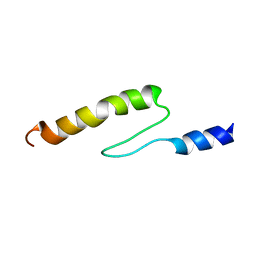 | | Solution structure of actin-binding domain of troponin in Ca2+-bound state | | Descriptor: | Troponin I, fast skeletal muscle | | Authors: | Murakami, K, Yumoto, F, Ohki, S, Yasunaga, T, Tanokura, M, Wakabayashi, T. | | Deposit date: | 2004-03-22 | | Release date: | 2005-09-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Ca(2+)-regulated Muscle Relaxation at Interaction Sites of Troponin with Actin and Tropomyosin
J.Mol.Biol., 352, 2005
|
|
1VDI
 
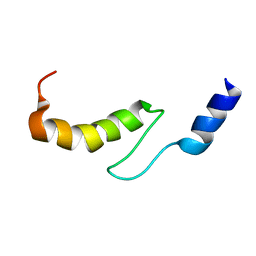 | | Solution structure of actin-binding domain of troponin in Ca2+-free state | | Descriptor: | Troponin I, fast skeletal muscle | | Authors: | Murakami, K, Yumoto, F, Ohki, S, Yasunaga, T, Tanokura, M, Wakabayashi, T. | | Deposit date: | 2004-03-22 | | Release date: | 2005-09-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Ca(2+)-regulated Muscle Relaxation at Interaction Sites of Troponin with Actin and Tropomyosin
J.Mol.Biol., 352, 2005
|
|
7DOD
 
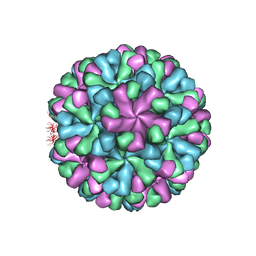 | | Capsid structure of human sapovirus | | Descriptor: | Calicivirin | | Authors: | Miyazaki, N, Murakami, K, Oka, T, Iwasaki, K, Katayama, K, Murata, K. | | Deposit date: | 2020-12-14 | | Release date: | 2021-12-15 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Atomic structure of human sapovirus capsid by single particle cryo-electron microscopy
To Be Published
|
|
6C9Y
 
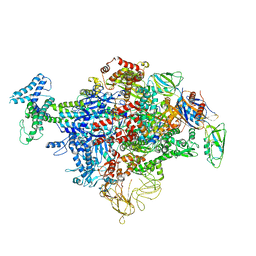 | | Cryo-EM structure of E. coli RNAP sigma70 holoenzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Narayanan, A, Vago, F, Li, K, Qayyum, M.Z, Yenool, D, Jiang, W, Murakami, K.S. | | Deposit date: | 2018-01-29 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Cryo-EM structure ofEscherichia colisigma70RNA polymerase and promoter DNA complex revealed a role of sigma non-conserved region during the open complex formation.
J. Biol. Chem., 293, 2018
|
|
6CA0
 
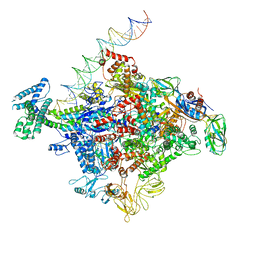 | | Cryo-EM structure of E. coli RNAP sigma70 open complex | | Descriptor: | DNA (35-MER), DNA (45-MER), DNA (5'-D(P*GP*CP*CP*GP*CP*GP*TP*CP*AP*GP*A)-3'), ... | | Authors: | Narayanan, A, Vago, F, Li, K, Qayyum, M.Z, Yernool, D, Jiang, W, Murakami, K.S. | | Deposit date: | 2018-01-29 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.75 Å) | | Cite: | Cryo-EM structure ofEscherichia colisigma70RNA polymerase and promoter DNA complex revealed a role of sigma non-conserved region during the open complex formation.
J. Biol. Chem., 293, 2018
|
|
6XI8
 
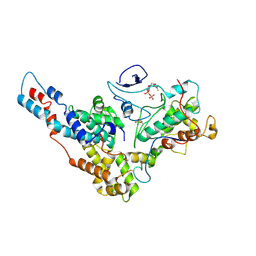 | | Yeast TFIIK (Kin28/Ccl1/Tfb3) Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Cyclin CCL1, ... | | Authors: | van Eeuwen, T, Murakami, K, Li, T, Tsai, K.L. | | Deposit date: | 2020-06-19 | | Release date: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structure of TFIIK for phosphorylation of CTD of RNA polymerase II.
Sci Adv, 7, 2021
|
|
5NWT
 
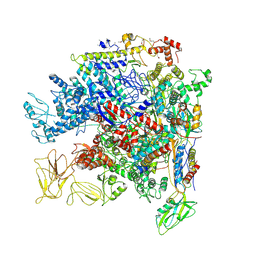 | | Crystal Structure of Escherichia coli RNA polymerase - Sigma54 Holoenzyme complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhang, X, Buck, M, Darbari, V.C, Yang, Y, Zhang, N, Lu, D, Glyde, R, Wang, Y, Winkelman, J, Gourse, R.L, Murakami, K.S. | | Deposit date: | 2017-05-08 | | Release date: | 2017-09-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.76 Å) | | Cite: | TRANSCRIPTION. Structures of the RNA polymerase-Sigma54 reveal new and conserved regulatory strategies.
Science, 349, 2015
|
|
6XI0
 
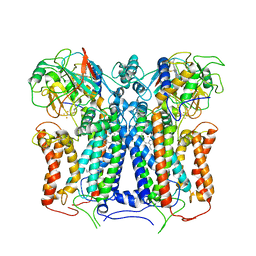 | | R. capsulatus cyt bc1 (CIII2) at 3.3A | | Descriptor: | Cytochrome b, Cytochrome c1, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Steimle, S, Van Eeuwen, T, Ozturk, Y, Kim, H.J, Braitbard, M, Selamoglu, N, Garcia, B.A, Schneidman-Duhovny, D, Murakami, K, Daldal, F. | | Deposit date: | 2020-06-19 | | Release date: | 2020-12-30 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of engineered active bc 1 -cbb 3 type CIII 2 CIV super-complexes and electronic communication between the complexes.
Nat Commun, 12, 2021
|
|
