5FMF
 
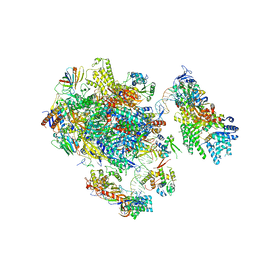 | | the P-lobe of RNA polymerase II pre-initiation complex | | Descriptor: | DNA REPAIR HELICASE RAD25, SSL2, DNA REPAIR HELICASE RAD3, ... | | Authors: | Murakami, K, Tsai, K, Kalisman, N, Bushnell, D.A, Asturias, F.J, Kornberg, R.D. | | Deposit date: | 2015-11-03 | | Release date: | 2015-11-25 | | Last modified: | 2017-07-12 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structure of an RNA Polymerase II Pre-Initiation Complex
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5XDR
 
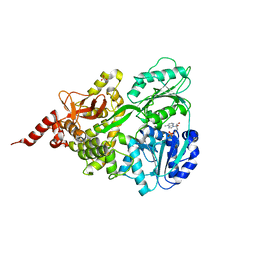 | | Crystal structure of human DEAH-box RNA helicase DHX15 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15, ... | | Authors: | Murakami, K, Nakano, K, Shimizu, T, Ohto, U. | | Deposit date: | 2017-03-29 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of human DEAH-box RNA helicase 15 reveals a domain organization of the mammalian DEAH/RHA family
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
2Z5H
 
 | | Crystal structure of the head-to-tail junction of tropomyosin complexed with a fragment of TnT | | Descriptor: | General control protein GCN4 and Tropomyosin alpha-1 chain, Tropomyosin alpha-1 chain and General control protein GCN4, Troponin T, ... | | Authors: | Murakami, K, Nozawa, K, Tomii, K, Kudou, N, Igarashi, N, Shirakihara, Y, Wakatsuki, S, Stewart, M, Yasunaga, T, Wakabayashi, T. | | Deposit date: | 2007-07-12 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural basis for tropomyosin overlap in thin (actin) filaments and the generation of a molecular swivel by troponin-T
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2Z5I
 
 | | Crystal structure of the head-to-tail junction of tropomyosin | | Descriptor: | General control protein GCN4 and Tropomyosin alpha-1 chain, MAGNESIUM ION, Tropomyosin alpha-1 chain and General control protein GCN4 | | Authors: | Murakami, K, Nozawa, K, Tomii, K, Kudou, N, Igarashi, N, Shirakihara, Y, Wakatsuki, S, Stewart, M, Yasunaga, T, Wakabayashi, T. | | Deposit date: | 2007-07-12 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for tropomyosin overlap in thin (actin) filaments and the generation of a molecular swivel by troponin-T
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
1VDI
 
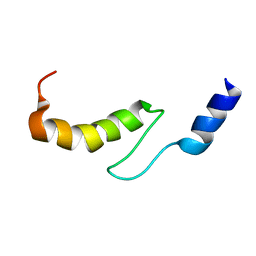 | | Solution structure of actin-binding domain of troponin in Ca2+-free state | | Descriptor: | Troponin I, fast skeletal muscle | | Authors: | Murakami, K, Yumoto, F, Ohki, S, Yasunaga, T, Tanokura, M, Wakabayashi, T. | | Deposit date: | 2004-03-22 | | Release date: | 2005-09-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Ca(2+)-regulated Muscle Relaxation at Interaction Sites of Troponin with Actin and Tropomyosin
J.Mol.Biol., 352, 2005
|
|
1VDJ
 
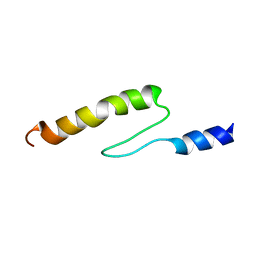 | | Solution structure of actin-binding domain of troponin in Ca2+-bound state | | Descriptor: | Troponin I, fast skeletal muscle | | Authors: | Murakami, K, Yumoto, F, Ohki, S, Yasunaga, T, Tanokura, M, Wakabayashi, T. | | Deposit date: | 2004-03-22 | | Release date: | 2005-09-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Ca(2+)-regulated Muscle Relaxation at Interaction Sites of Troponin with Actin and Tropomyosin
J.Mol.Biol., 352, 2005
|
|
3A5M
 
 | | Crystal Structure of a Dictyostelium P109I Mg2+-Actin in Complex with Human Gelsolin Segment 1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Gelsolin, ... | | Authors: | Murakami, K, Yasunaga, T, Noguchi, T.Q, Uyeda, T.Q, Wakabayashi, T. | | Deposit date: | 2009-08-09 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for actin assembly, activation of ATP hydrolysis, and delayed phosphate release
Cell(Cambridge,Mass.), 143, 2010
|
|
3A5L
 
 | | Crystal Structure of a Dictyostelium P109A Mg2+-Actin in Complex with Human Gelsolin Segment 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Gelsolin, ... | | Authors: | Murakami, K, Yasunaga, T, Noguchi, T.Q, Uyeda, T.Q, Wakabayashi, T. | | Deposit date: | 2009-08-09 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for actin assembly, activation of ATP hydrolysis, and delayed phosphate release
Cell(Cambridge,Mass.), 143, 2010
|
|
3A5N
 
 | | Crystal Structure of a Dictyostelium P109A Ca2+-Actin in Complex with Human Gelsolin Segment 1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Gelsolin, ... | | Authors: | Murakami, K, Yasunaga, T, Noguchi, T.Q, Uyeda, T.Q, Wakabayashi, T. | | Deposit date: | 2009-08-09 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural basis for actin assembly, activation of ATP hydrolysis, and delayed phosphate release
Cell(Cambridge,Mass.), 143, 2010
|
|
3A5O
 
 | | Crystal Structure of a Dictyostelium P109I Ca2+-Actin in Complex with Human Gelsolin Segment 1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Gelsolin, ... | | Authors: | Murakami, K, Yasunaga, T, Noguchi, T.Q, Uyeda, T.Q, Wakabayashi, T. | | Deposit date: | 2009-08-09 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for actin assembly, activation of ATP hydrolysis, and delayed phosphate release
Cell(Cambridge,Mass.), 143, 2010
|
|
7DOD
 
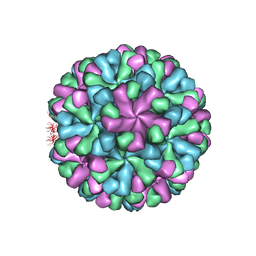 | | Capsid structure of human sapovirus | | Descriptor: | Calicivirin | | Authors: | Miyazaki, N, Murakami, K, Oka, T, Iwasaki, K, Katayama, K, Murata, K. | | Deposit date: | 2020-12-14 | | Release date: | 2021-12-15 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Atomic structure of human sapovirus capsid by single particle cryo-electron microscopy
To Be Published
|
|
3G37
 
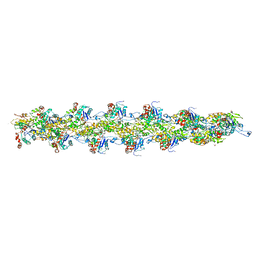 | | Cryo-EM structure of actin filament in the presence of phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Wakabayshi, T, Murakami, K, Yasunaga, T, Noguchi, T.Q, Uyeda, T.Q. | | Deposit date: | 2009-02-02 | | Release date: | 2010-11-03 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structural basis for actin assembly, activation of ATP hydrolysis, and delayed phosphate release
Cell(Cambridge,Mass.), 143, 2010
|
|
7ML0
 
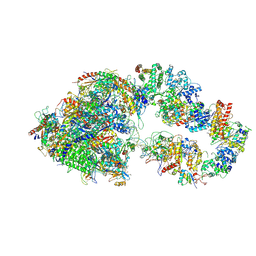 | | RNA polymerase II pre-initiation complex (PIC1) | | Descriptor: | BJ4_G0050160.mRNA.1.CDS.1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Fujiwara, R, Kim, H.J, Gorbea Colon, J.J, Steimle, S, Garcia, B.A, Murakami, K. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7MEI
 
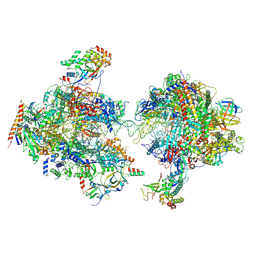 | | Composite structure of EC+EC | | Descriptor: | DNA (74-MER), DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Murakami, K. | | Deposit date: | 2021-04-06 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7MKA
 
 | | Structure of EC+EC (leading EC-focused) | | Descriptor: | DNA (40-MER), DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Murakami, K. | | Deposit date: | 2021-04-22 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7MK9
 
 | | Complex structure of trailing EC of EC+EC (trailing EC-focused) | | Descriptor: | DNA (40-MER), DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Murakami, K. | | Deposit date: | 2021-04-22 | | Release date: | 2022-04-27 | | Last modified: | 2023-05-17 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7ML1
 
 | | RNA polymerase II pre-initiation complex (PIC2) | | Descriptor: | BJ4_G0050160.mRNA.1.CDS.1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Fujiwara, R, Kim, H.J, Gorbea Colon, J.J, Steimle, S, Garcia, B.A, Murakami, K. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7ML3
 
 | | General transcription factor TFIIH (weak binding) | | Descriptor: | BJ4_G0050160.mRNA.1.CDS.1, General transcription and DNA repair factor IIH, General transcription and DNA repair factor IIH helicase subunit XPB, ... | | Authors: | Yang, C, Fujiwara, R, Kim, H.J, Gorbea Colon, J.J, Steimle, S, Garcia, B.A, Murakami, K. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7ML4
 
 | | RNA polymerase II initially transcribing complex (ITC) | | Descriptor: | BJ4_G0050160.mRNA.1.CDS.1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Fujiwara, R, Kim, H.J, Gorbea Colon, J.J, Steimle, S, Garcia, B.A, Murakami, K. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-02 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7ML2
 
 | | RNA polymerase II pre-initiation complex (PIC3) | | Descriptor: | BJ4_G0004860.mRNA.1.CDS.1, BJ4_G0050160.mRNA.1.CDS.1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Yang, C, Fujiwara, R, Kim, H.J, Gorbea Colon, J.J, Steimle, S, Garcia, B.A, Murakami, K. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
8UOQ
 
 | | Composite map of PIC_delta_TFIIK form2 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Yang, C, Murakami, K. | | Deposit date: | 2023-10-20 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Transcription start site scanning requires the fungi-specific hydrophobic loop of Tfb3.
Nucleic Acids Res., 2024
|
|
8UOT
 
 | | Composite map of PICdeltaTFIIK form1 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Yang, C, Murakami, K. | | Deposit date: | 2023-10-20 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Transcription start site scanning requires the fungi-specific hydrophobic loop of Tfb3.
Nucleic Acids Res., 2024
|
|
8UMH
 
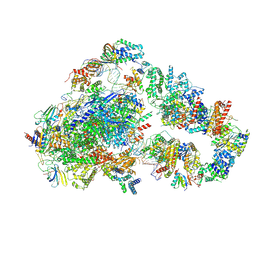 | | Consensus map of PICdeltaTFIIK form2 | | Descriptor: | DNA (63-MER), DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Murakami, K. | | Deposit date: | 2023-10-17 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | consensus map of PICdeltaTFIIK form2
To be published
|
|
8UMI
 
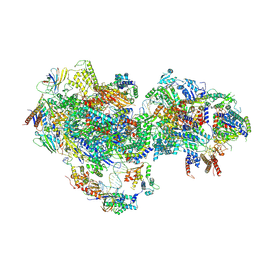 | | consensus map of PICdeltaTFIIK form1 | | Descriptor: | DNA (64-MER), DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Murakami, K. | | Deposit date: | 2023-10-17 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | consensus map of PICdeltaTFIIK form1
To be published
|
|
7M2U
 
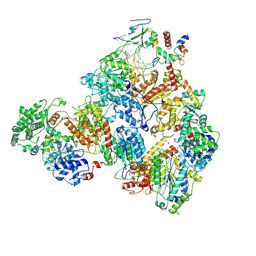 | | Nucleotide Excision Repair complex TFIIH Rad4-33 | | Descriptor: | CALCIUM ION, DNA repair helicase RAD25, DNA repair helicase RAD3, ... | | Authors: | van Eeuwen, T, Murakami, K. | | Deposit date: | 2021-03-17 | | Release date: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Cryo-EM structure of TFIIH/Rad4-Rad23-Rad33 in damaged DNA opening in nucleotide excision repair.
Nat Commun, 12, 2021
|
|
