7X7K
 
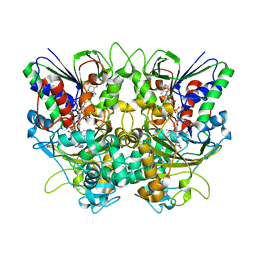 | | Ancestral L-Lys oxidase (AncLLysO-2) L-Arg binding form | | Descriptor: | ARGININE, FAD dependent enzyme, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Motoyama, T, Ishida, C, Hasebe, F, Ito, S, Nakano, S. | | Deposit date: | 2022-03-09 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Reaction Mechanism of Ancestral l-Lys alpha-Oxidase from Caulobacter Species Studied by Biochemical, Structural, and Computational Analysis
Acs Omega, 7, 2022
|
|
7X7J
 
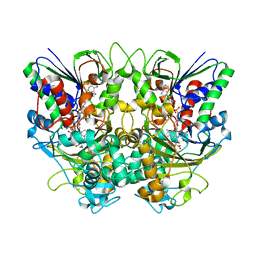 | | Ancestral L-Lys oxidase (AncLLysO-2) L-Lys binding form | | Descriptor: | FAD dependent enzyme, FLAVIN-ADENINE DINUCLEOTIDE, LYSINE | | Authors: | Motoyama, T, Ishida, C, Hasebe, F, Ito, S, Nakano, S. | | Deposit date: | 2022-03-09 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Reaction Mechanism of Ancestral l-Lys alpha-Oxidase from Caulobacter Species Studied by Biochemical, Structural, and Computational Analysis
Acs Omega, 7, 2022
|
|
7X7I
 
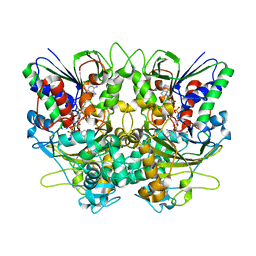 | | Ancestral L-Lys oxidase (AncLLysO-2) ligand free form | | Descriptor: | FAD dependent enzyme, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Motoyama, T, Ishida, C, Hasebe, F, Ito, S, Nakano, S. | | Deposit date: | 2022-03-09 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Reaction Mechanism of Ancestral l-Lys alpha-Oxidase from Caulobacter Species Studied by Biochemical, Structural, and Computational Analysis
Acs Omega, 7, 2022
|
|
7BXS
 
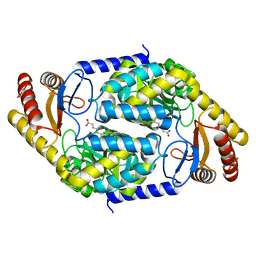 | | 2-amino-3-ketobutyrate CoA ligase from Cupriavidus necator glycine binding form | | Descriptor: | 2-amino-3-ketobutyrate coenzyme A ligase, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE] | | Authors: | Motoyama, T, Nakano, S, Hasebe, F, Miyoshi, N, Ito, S. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Chemoenzymatic synthesis of 3-ethyl-2,5-dimethylpyrazine by L-threonine 3-dehydrogenase and 2-amino-3-ketobutyrate CoA ligase/L-threonine aldolase
Commun Chem, 4, 2021
|
|
7BXQ
 
 | | 2-amino-3-ketobutyrate CoA ligase from Cupriavidus necator L-Threonine binding form | | Descriptor: | 2-amino-3-ketobutyrate coenzyme A ligase, N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-allothreonine | | Authors: | Motoyama, T, Nakano, S, Hasebe, F, Miyoshi, N, Ito, S. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Chemoenzymatic synthesis of 3-ethyl-2,5-dimethylpyrazine by L-threonine 3-dehydrogenase and 2-amino-3-ketobutyrate CoA ligase/L-threonine aldolase
Commun Chem, 4, 2021
|
|
7BXP
 
 | | 2-amino-3-ketobutyrate CoA ligase from Cupriavidus necator | | Descriptor: | 2-amino-3-ketobutyrate coenzyme A ligase | | Authors: | Motoyama, T, Nakano, S, Hasebe, F, Miyoshi, N, Ito, S. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Chemoenzymatic synthesis of 3-ethyl-2,5-dimethylpyrazine by L-threonine 3-dehydrogenase and 2-amino-3-ketobutyrate CoA ligase/L-threonine aldolase
Commun Chem, 4, 2021
|
|
7BXR
 
 | | 2-amino-3-ketobutyrate CoA ligase from Cupriavidus necator 3-Hydroxynorvaline binding form | | Descriptor: | (2S,3R)-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]-3-oxidanyl-pentanoic acid, 2-amino-3-ketobutyrate coenzyme A ligase | | Authors: | Motoyama, T, Nakano, S, Hasebe, F, Miyoshi, N, Ito, S. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Chemoenzymatic synthesis of 3-ethyl-2,5-dimethylpyrazine by L-threonine 3-dehydrogenase and 2-amino-3-ketobutyrate CoA ligase/L-threonine aldolase
Commun Chem, 4, 2021
|
|
5Y1F
 
 | | Monomeric L-threonine 3-dehydrogenase from metagenome database (NAD+ bound form) | | Descriptor: | NAD dependent epimerase/dehydratase family, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Motoyama, T, Nakano, S, Yamamoto, Y, Tokiwa, H, Asano, Y, Ito, S. | | Deposit date: | 2017-07-20 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Product Release Mechanism Associated with Structural Changes in Monomeric l-Threonine 3-Dehydrogenase.
Biochemistry, 56, 2017
|
|
5Y1G
 
 | | Monomeric L-threonine 3-dehydrogenase from metagenome database (AKB and NADH bound form) | | Descriptor: | 2-AMINO-3-KETOBUTYRIC ACID, NAD dependent epimerase/dehydratase family, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Motoyama, T, Nakano, S, Yamamoto, Y, Tokiwa, H, Asano, Y, Ito, S. | | Deposit date: | 2017-07-20 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Product Release Mechanism Associated with Structural Changes in Monomeric l-Threonine 3-Dehydrogenase.
Biochemistry, 56, 2017
|
|
5Y1E
 
 | | monomeric L-threonine 3-dehydrogenase from metagenome database (L-Ser and NAD+ bound form) | | Descriptor: | NAD dependent epimerase/dehydratase family, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SERINE | | Authors: | Motoyama, T, Nakano, S, Yamamoto, Y, Tokiwa, H, Asano, Y, Ito, S. | | Deposit date: | 2017-07-20 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Product Release Mechanism Associated with Structural Changes in Monomeric l-Threonine 3-Dehydrogenase.
Biochemistry, 56, 2017
|
|
5Y1D
 
 | | Monomeric L-threonine 3-dehydrogenase from metagenome database (apo form) | | Descriptor: | NAD dependent epimerase/dehydratase family | | Authors: | Motoyama, T, Nakano, S, Yamamoto, Y, Tokiwa, H, Asano, Y, Ito, S. | | Deposit date: | 2017-07-20 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Product Release Mechanism Associated with Structural Changes in Monomeric l-Threonine 3-Dehydrogenase.
Biochemistry, 56, 2017
|
|
7CGV
 
 | | Full consensus L-threonine 3-dehydrogenase, FcTDH-IIYM (NAD+ bound form) | | Descriptor: | Artificial L-threonine 3-dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Motoyama, T, Hiramatsu, N, Asano, Y, Nakano, S, Ito, S. | | Deposit date: | 2020-07-02 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Protein Sequence Selection Method That Enables Full Consensus Design of Artificial l-Threonine 3-Dehydrogenases with Unique Enzymatic Properties.
Biochemistry, 59, 2020
|
|
1IDP
 
 | |
3WQH
 
 | | Crystal Structure of human DPP-IV in complex with Anagliptin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, N-[2-({2-[(2S)-2-cyanopyrrolidin-1-yl]-2-oxoethyl}amino)-2-methylpropyl]-2-methylpyrazolo[1,5-a]pyrimidine-6-carboxamide | | Authors: | Watanabe, Y.S, Okada, S, Motoyama, T, Takahashi, R, Adachi, H, Oka, M. | | Deposit date: | 2014-01-27 | | Release date: | 2015-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Anagliptin, a potent dipeptidyl peptidase IV inhibitor: its single-crystal structure and enzyme interactions.
J Enzyme Inhib Med Chem, 30, 2015
|
|
2STD
 
 | | SCYTALONE DEHYDRATASE COMPLEXED WITH TIGHT-BINDING INHIBITOR CARPROPAMID | | Descriptor: | ((1RS,3SR)-2,2-DICHLORO-N-[(R)-1-(4-CHLOROPHENYL)ETHYL]-1-ETHYL-3-METHYLCYCLOPROPANECARBOXAMIDE, SCYTALONE DEHYDRATASE, SULFATE ION | | Authors: | Nakasako, M, Motoyama, T, Kurahashi, Y, Yamaguchi, I. | | Deposit date: | 1997-12-21 | | Release date: | 1999-02-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cryogenic X-ray crystal structure analysis for the complex of scytalone dehydratase of a rice blast fungus and its tight-binding inhibitor, carpropamid: the structural basis of tight-binding inhibition.
Biochemistry, 37, 1998
|
|
6KOG
 
 | | Ketosynthase domain in tenuazonic acid synthetase 1 (TAS1). | | Descriptor: | GLYCEROL, Hybrid PKS-NRPS synthetase TAS1, SULFATE ION | | Authors: | Yun, C.S, Nishimoto, K, Motoyama, T, Hino, T, Nagano, S, Osada, H. | | Deposit date: | 2019-08-10 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Unique features of the ketosynthase domain in a nonribosomal peptide synthetase-polyketide synthase hybrid enzyme, tenuazonic acid synthetase 1.
J.Biol.Chem., 295, 2020
|
|
6JNO
 
 | | RXRa structure complexed with CU-6PMN | | Descriptor: | 7-oxidanyl-2-oxidanylidene-6-(3,5,5,8,8-pentamethyl-6,7-dihydronaphthalen-2-yl)chromene-3-carboxylic acid, Retinoic acid receptor RXR-alpha | | Authors: | Kawasaki, M, Nakano, S, Motoyama, T, Yamada, S, Watanabe, M, Takamura, Y, Fujihara, M, Tokiwa, H, Kakuta, H, Ito, S. | | Deposit date: | 2019-03-17 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Competitive Binding Assay with an Umbelliferone-Based Fluorescent Rexinoid for Retinoid X Receptor Ligand Screening.
J.Med.Chem., 62, 2019
|
|
6JNR
 
 | | RXRa structure complexed with CU-6PMN and SRC1 peptide. | | Descriptor: | 7-oxidanyl-2-oxidanylidene-6-(3,5,5,8,8-pentamethyl-6,7-dihydronaphthalen-2-yl)chromene-3-carboxylic acid, HIS-LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN, Retinoic acid receptor RXR-alpha | | Authors: | Kawasaki, M, Nakano, S, Motoyama, T, Yamada, S, Watanabe, M, Takamura, Y, Fujihara, M, Tokiwa, H, Kakuta, H, Ito, S. | | Deposit date: | 2019-03-18 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | RXRa structure complexed with CU-6PMN and SRC1 peptide.
To Be Published
|
|
5Z75
 
 | | Artificial L-threonine 3-dehydrogenase designed by ancestral sequence reconstruction. | | Descriptor: | Artificial L-threonine 3-dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, NONAETHYLENE GLYCOL, ... | | Authors: | Nakano, S, Motoyama, T, Miyashita, Y, Ishizuka, Y, Matsuo, N, Tokiwa, H, Shinoda, S, Asano, Y, Ito, S. | | Deposit date: | 2018-01-27 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Benchmark Analysis of Native and Artificial NAD+-Dependent Enzymes Generated by a Sequence-Based Design Method with or without Phylogenetic Data.
Biochemistry, 57, 2018
|
|
5Z76
 
 | | Artificial L-threonine 3-dehydrogenase designed by full consensus design | | Descriptor: | Artificial L-threonine 3-dehydrogenase | | Authors: | Nakano, S, Motoyama, T, Miyashita, Y, Ishizuka, Y, Matsuo, N, Tokiwa, H, Shinoda, S, Asano, Y, Ito, S. | | Deposit date: | 2018-01-27 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Benchmark Analysis of Native and Artificial NAD+-Dependent Enzymes Generated by a Sequence-Based Design Method with or without Phylogenetic Data.
Biochemistry, 57, 2018
|
|
7CFO
 
 | | Crystal structure of human RXRalpha ligand binding domain complexed with CBTF-EE. | | Descriptor: | 1-[3-(2-ethoxyethoxy)-5,5,8,8-tetramethyl-6,7-dihydronaphthalen-2-yl]-2-(trifluoromethyl)benzimidazole-5-carboxylic acid, GLYCEROL, Retinoic acid receptor RXR-alpha | | Authors: | Watanabe, M, Fujihara, M, Motoyama, T, Kawasaki, M, Yamada, S, Takamura, Y, Ito, S, Makishima, M, Nakano, S, Kakuta, H. | | Deposit date: | 2020-06-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of a "Gatekeeper" Antagonist that Blocks Entry Pathway to Retinoid X Receptors (RXRs) without Allosteric Ligand Inhibition in Permissive RXR Heterodimers.
J.Med.Chem., 64, 2021
|
|
