3WD8
 
 | | TypeIII polyketide synthases | | Descriptor: | GLYCEROL, Type III polyketide synthase quinolone synthase | | Authors: | Mori, T, Shimokawa, Y, Matsui, T, Morita, H, Abe, I. | | Deposit date: | 2013-06-10 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.463 Å) | | Cite: | Cloning, characterization, and crystal structure analysis of novel type III polyketide synthases from Citrus microcarpa
To be Published
|
|
3WXZ
 
 | | The structure of the I375F mutant of CsyB | | Descriptor: | Putative uncharacterized protein csyB | | Authors: | Mori, T, Yang, D, Matsui, T, Morita, H, Fujii, I, Abe, I. | | Deposit date: | 2014-08-13 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Structural basis for the formation of acylalkylpyrones from two beta-ketoacyl units by the fungal type III polyketide synthase CsyB.
J.Biol.Chem., 290, 2015
|
|
3WY0
 
 | | The I375W mutant of CsyB complexed with CoA-SH | | Descriptor: | COENZYME A, Putative uncharacterized protein csyB | | Authors: | Mori, T, Yang, D, Matsui, T, Morita, H, Fujii, I, Abe, I. | | Deposit date: | 2014-08-13 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis for the formation of acylalkylpyrones from two beta-ketoacyl units by the fungal type III polyketide synthase CsyB.
J.Biol.Chem., 290, 2015
|
|
3WXY
 
 | | Crystal structure of CsyB complexed with CoA-SH | | Descriptor: | COENZYME A, Putative uncharacterized protein csyB | | Authors: | Mori, T, Yang, D, Matsui, T, Morita, H, Fujii, I, Abe, I. | | Deposit date: | 2014-08-13 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.706 Å) | | Cite: | Structural basis for the formation of acylalkylpyrones from two beta-ketoacyl units by the fungal type III polyketide synthase CsyB.
J.Biol.Chem., 290, 2015
|
|
3X27
 
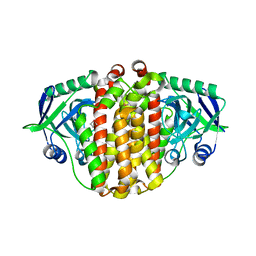 | | Structure of McbB in complex with tryptophan | | Descriptor: | Cucumopine synthase, TRYPTOPHAN | | Authors: | Mori, T, Sahashi, S, Morita, H, Abe, I. | | Deposit date: | 2014-12-10 | | Release date: | 2015-10-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.481 Å) | | Cite: | Structural Basis for beta-Carboline Alkaloid Production by the Microbial Homodimeric Enzyme McbB
Chem.Biol., 22, 2015
|
|
8KDL
 
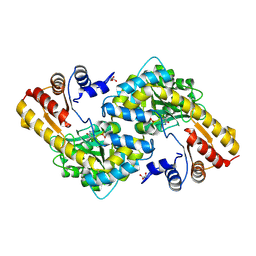 | |
8KDK
 
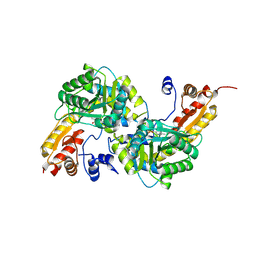 | |
7YN3
 
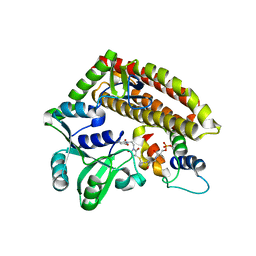 | | Crystal structure of CcbD complex with CcbZ carrier protein domain | | Descriptor: | 1,1'-ethane-1,2-diyldipyrrolidine-2,5-dione, 4'-PHOSPHOPANTETHEINE, CcbD, ... | | Authors: | Mori, T, Lyu, S, Abe, I. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of CcbD complex with carrier protein
To Be Published
|
|
7YN1
 
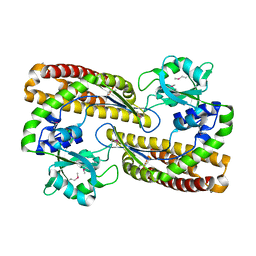 | |
7YN2
 
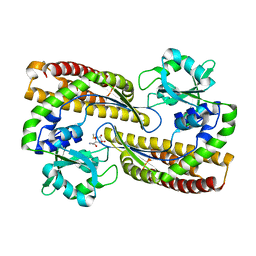 | | Crystal structure of CcbD with methylthiolincosamide | | Descriptor: | (2R,3R,4S,5R,6R)-2-[(1R,2R)-1-azanyl-2-oxidanyl-propyl]-6-methylsulfanyl-oxane-3,4,5-triol, CcbD | | Authors: | Mori, T, Lyu, S, Abe, I. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of CcbD with methylthiolincosamide
To Be Published
|
|
7DRE
 
 | | Cryo-EM structure of DfgA-B at 2.54 angstrom resolution | | Descriptor: | DfgB, Sugar phosphate isomerase/epimerase | | Authors: | Mori, T, Moriya, T, Adachi, N, Senda, T, Abe, I. | | Deposit date: | 2020-12-28 | | Release date: | 2021-12-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
7DRD
 
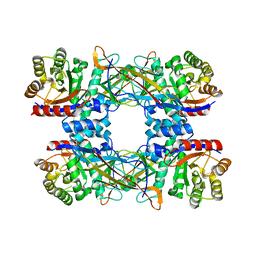 | | Cryo-EM structure of DgpB-C at 2.85 angstrom resolution | | Descriptor: | AP_endonuc_2 domain-containing protein, DgpB | | Authors: | Mori, T, Moriya, T, Adachi, N, Senda, T, Abe, I. | | Deposit date: | 2020-12-28 | | Release date: | 2021-12-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
7WIJ
 
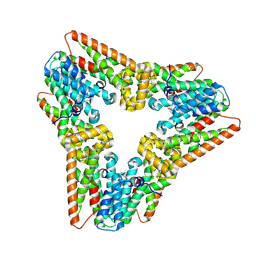 | |
7WPY
 
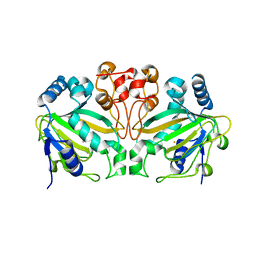 | | AndA_M119A_N121V variant | | Descriptor: | Dioxygenase andA, FE (III) ION | | Authors: | Mori, T, Chen, H, Abe, I. | | Deposit date: | 2022-01-24 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | AndA_M119A_N121V variant
To Be Published
|
|
7EXZ
 
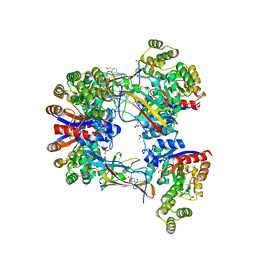 | | DgpB-DgpC complex apo 2.5 angstrom | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AP_endonuc_2 domain-containing protein, DgpB, ... | | Authors: | Mori, T, Senda, M, Senda, T, Abe, I. | | Deposit date: | 2021-05-29 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
7EXB
 
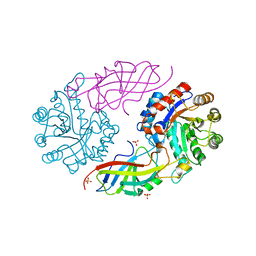 | | DfgA-DfgB complex apo 2.4 angstrom | | Descriptor: | DfgB, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Mori, T, Senda, M, Senda, T, Abe, I. | | Deposit date: | 2021-05-26 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
2ZPY
 
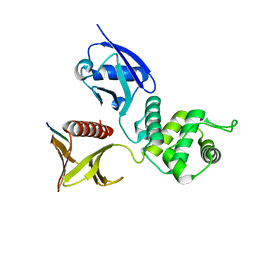 | | Crystal structure of the mouse radxin FERM domain complexed with the mouse CD44 cytoplasmic peptide | | Descriptor: | CD44 antigen, Radixin | | Authors: | Mori, T, Kitano, K, Terawaki, S, Maesaki, R, Fukami, Y, Hakoshima, T. | | Deposit date: | 2008-07-31 | | Release date: | 2008-08-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for CD44 recognition by ERM proteins
J.Biol.Chem., 283, 2008
|
|
7ENB
 
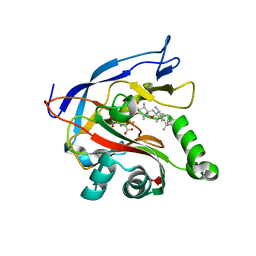 | |
7EMZ
 
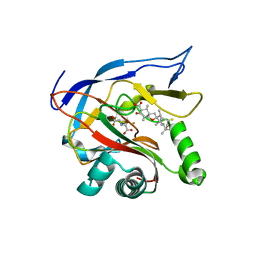 | |
5YIZ
 
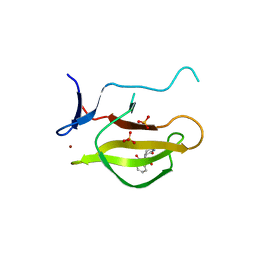 | |
5YJ0
 
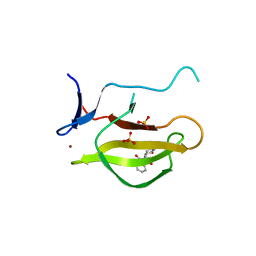 | |
5YJ1
 
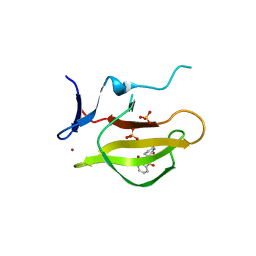 | | Mouse Cereblon thalidomide binding domain complexed with R-form thalidomide | | Descriptor: | 2-[(3~{R})-2,6-bis(oxidanylidene)piperidin-3-yl]isoindole-1,3-dione, Protein cereblon, SULFATE ION, ... | | Authors: | Mori, T, Hakoshima, T. | | Deposit date: | 2017-10-06 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of thalidomide enantiomer binding to cereblon
Sci Rep, 8, 2018
|
|
5X9J
 
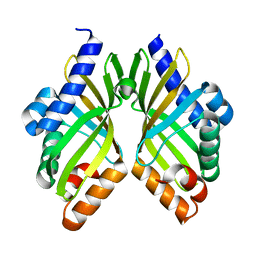 | |
3WX1
 
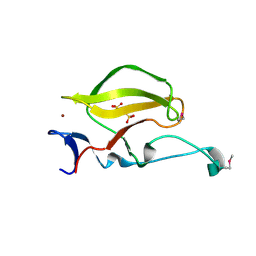 | | Mouse Cereblon thalidomide binding domain, selenomethionine derivative | | Descriptor: | Protein cereblon, SULFATE ION, ZINC ION | | Authors: | Mori, T, Ito, T, Hirano, Y, Yamaguchi, Y, Handa, H, Hakoshima, T. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of the human Cereblon-DDB1-lenalidomide complex reveals basis for responsiveness to thalidomide analogs
Nat.Struct.Mol.Biol., 21, 2014
|
|
3WX2
 
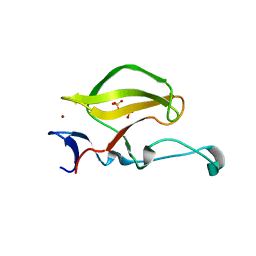 | | Mouse Cereblon thalidomide binding domain, native | | Descriptor: | Protein cereblon, SULFATE ION, ZINC ION | | Authors: | Mori, T, Ito, T, Hirano, Y, Yamaguchi, Y, Handa, H, Hakoshima, T. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the human Cereblon-DDB1-lenalidomide complex reveals basis for responsiveness to thalidomide analogs
Nat.Struct.Mol.Biol., 21, 2014
|
|
