8GQ9
 
 | | Crystal structure of lasso peptide epimerase MslH | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Nakashima, Y, Morita, H. | | Deposit date: | 2022-08-29 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of lasso peptide epimerase MslH reveals metal-dependent acid/base catalytic mechanism.
Nat Commun, 14, 2023
|
|
8GQB
 
 | |
8GQA
 
 | |
5F9K
 
 | | Dictyostelium discoideum dUTPase at 2.2 Angstrom | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, MAGNESIUM ION, ... | | Authors: | Inoguchi, N, Chia, C.P, Heck, K, Moriyama, H. | | Deposit date: | 2015-12-09 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.179 Å) | | Cite: | Mitochondrial localization of Dictyostelium discoideum dUTPase mediated by its N-terminus.
Bmc Res Notes, 13, 2020
|
|
5GJI
 
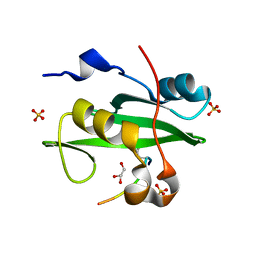 | | PI3K p85 N-terminal SH2 domain/CD28-derived peptide complex | | Descriptor: | GLYCEROL, Phosphatidylinositol 3-kinase regulatory subunit alpha, SULFATE ION, ... | | Authors: | Inaba, S, Numoto, N, Morii, H, Ogawa, S, Ikura, T, Abe, R, Ito, N, Oda, M. | | Deposit date: | 2016-06-30 | | Release date: | 2016-12-14 | | Last modified: | 2017-05-10 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Crystal Structures and Thermodynamic Analysis Reveal Distinct Mechanisms of CD28 Phosphopeptide Binding to the Src Homology 2 (SH2) Domains of Three Adaptor Proteins
J. Biol. Chem., 292, 2017
|
|
5GJH
 
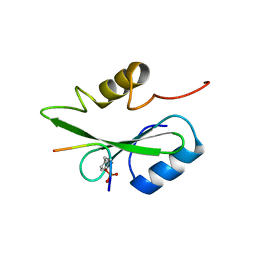 | | Gads SH2 domain/CD28-derived peptide complex | | Descriptor: | GRB2-related adapter protein 2, T-cell-specific surface glycoprotein CD28 | | Authors: | Inaba, S, Numoto, N, Morii, H, Ogawa, S, Ikura, T, Abe, R, Ito, N, Oda, M. | | Deposit date: | 2016-06-30 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structures and Thermodynamic Analysis Reveal Distinct Mechanisms of CD28 Phosphopeptide Binding to the Src Homology 2 (SH2) Domains of Three Adaptor Proteins
J. Biol. Chem., 292, 2017
|
|
1EID
 
 | |
1WLS
 
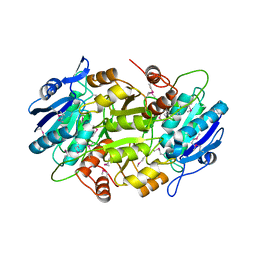 | | Crystal structure of L-asparaginase I homologue protein from Pyrococcus horikoshii | | Descriptor: | L-asparaginase | | Authors: | Yao, M, Morita, H, Yasutake, Y, Tanaka, I. | | Deposit date: | 2004-06-29 | | Release date: | 2005-03-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure of the type I L-asparaginase from the hyperthermophilic archaeon Pyrococcus horikoshii at 2.16 angstroms resolution.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1EIC
 
 | |
1WPW
 
 | | Crystal Structure of IPMDH from Sulfolobus tokodaii | | Descriptor: | 3-isopropylmalate dehydrogenase, MAGNESIUM ION | | Authors: | Hirose, R, Sakurai, M, Suzuki, T, Moriyama, H, Sato, T, Yamagishi, A, Oshima, T, Tanaka, N. | | Deposit date: | 2004-09-14 | | Release date: | 2004-10-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of IPMDH from Sulfolobus tokodaii
To be Published
|
|
4OOQ
 
 | | apo-dUTPase from Arabidopsis thaliana | | Descriptor: | Deoxyuridine 5'-triphosphate nucleotidohydrolase, MAGNESIUM ION, SULFATE ION | | Authors: | Inoguchi, N, Bajaj, M, Moriyama, H. | | Deposit date: | 2014-02-03 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Structural insights into the mechanism defining substrate affinity in Arabidopsis thaliana dUTPase: the role of tryptophan 93 in ligand orientation.
BMC Res Notes, 8, 2015
|
|
2ZVK
 
 | | Crystal structure of PCNA in complex with DNA polymerase eta fragment | | Descriptor: | DNA polymerase eta, Proliferating cell nuclear antigen | | Authors: | Hishiki, A, Hashimoto, H, Hanafusa, T, Kamei, K, Ohashi, E, Shimizu, T, Ohmori, H, Sato, M. | | Deposit date: | 2008-11-11 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Novel Interactions between Human Translesion Synthesis Polymerases and Proliferating Cell Nuclear Antigen
J.Biol.Chem., 284, 2009
|
|
1TZ4
 
 | | [hPP19-23]-pNPY bound to DPC Micelles | | Descriptor: | neuropeptide Y,Pancreatic prohormone,neuropeptide Y | | Authors: | Lerch, M, Kamimori, H, Folkers, G, Aguilar, M.I, Beck-Sickinger, A.G, Zerbe, O. | | Deposit date: | 2004-07-09 | | Release date: | 2005-07-05 | | Last modified: | 2020-01-01 | | Method: | SOLUTION NMR | | Cite: | Strongly Altered Receptor Binding Properties in PP and NPY Chimeras Are Accompanied by Changes
in Structure and Membrane Binding
Biochemistry, 44, 2005
|
|
2ZVM
 
 | | Crystal structure of PCNA in complex with DNA polymerase iota fragment | | Descriptor: | DNA polymerase iota, Proliferating cell nuclear antigen | | Authors: | Hishiki, A, Hashimoto, H, Hanafusa, T, Kamei, K, Ohashi, E, Shimizu, T, Ohmori, H, Sato, M. | | Deposit date: | 2008-11-11 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Novel Interactions between Human Translesion Synthesis Polymerases and Proliferating Cell Nuclear Antigen
J.Biol.Chem., 284, 2009
|
|
6ICG
 
 | | Grb2 SH2 domain in phosphopeptide free form | | Descriptor: | GLYCEROL, Growth factor receptor-bound protein 2, SULFATE ION | | Authors: | Hosoe, Y, Numoto, N, Inaba, S, Ogawa, S, Morii, H, Abe, R, Ito, N, Oda, M. | | Deposit date: | 2018-09-06 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural and functional properties of Grb2 SH2 dimer in CD28 binding.
Biophys Physicobio., 16, 2019
|
|
7W6F
 
 | | Polyketide cyclase OAC-F24I mutant from Cannabis sativa in complex with 6-nonylresorcylic acid | | Descriptor: | 2-nonyl-4,6-bis(oxidanyl)benzoic acid, GLYCEROL, Olivetolic acid cyclase | | Authors: | Nakashima, Y, Lee, Y.E, Morita, H. | | Deposit date: | 2021-12-01 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Dual Engineering of Olivetolic Acid Cyclase and Tetraketide Synthase to Generate Longer Alkyl-Chain Olivetolic Acid Analogs.
Org.Lett., 24, 2022
|
|
7W6G
 
 | | TKS-L190G mutant from Cannabis sativa in complex with lauroyl-CoA | | Descriptor: | 3,5,7-trioxododecanoyl-CoA synthase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Nakashima, Y, Lee, Y.E, Morita, H. | | Deposit date: | 2021-12-01 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dual Engineering of Olivetolic Acid Cyclase and Tetraketide Synthase to Generate Longer Alkyl-Chain Olivetolic Acid Analogs.
Org.Lett., 24, 2022
|
|
7W6E
 
 | |
7W6D
 
 | |
1VDZ
 
 | | Crystal structure of A-type ATPase catalytic subunit A from Pyrococcus horikoshii OT3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, A-type ATPase subunit A | | Authors: | Maegawa, Y, Morita, H, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2004-03-26 | | Release date: | 2005-06-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of A-type ATPase catalytic subunit A from Pyrococcus horikoshii OT3
To be Published
|
|
1FS3
 
 | |
2ZVL
 
 | | Crystal structure of PCNA in complex with DNA polymerase kappa fragment | | Descriptor: | DNA polymerase kappa, Proliferating cell nuclear antigen, SULFATE ION, ... | | Authors: | Hishiki, A, Hashimoto, H, Hanafusa, T, Kamei, K, Ohashi, E, Shimizu, T, Ohmori, H, Sato, M. | | Deposit date: | 2008-11-11 | | Release date: | 2009-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Novel Interactions between Human Translesion Synthesis Polymerases and Proliferating Cell Nuclear Antigen
J.Biol.Chem., 284, 2009
|
|
1TZ5
 
 | | [pNPY19-23]-hPP bound to DPC Micelles | | Descriptor: | Pancreatic prohormone,neuropeptide Y,Pancreatic prohormone | | Authors: | Lerch, M, Kamimori, H, Folkers, G, Aguilar, M.I, Beck-Sickinger, A.G, Zerbe, O. | | Deposit date: | 2004-07-09 | | Release date: | 2005-07-05 | | Last modified: | 2020-01-01 | | Method: | SOLUTION NMR | | Cite: | Strongly Altered Receptor Binding Properties in PP and NPY Chimeras Are Accompanied by Changes
in Structure and Membrane Binding
Biochemistry, 44, 2005
|
|
1G2U
 
 | | THE STRUCTURE OF THE MUTANT, A172V, OF 3-ISOPROPYLMALATE DEHYDROGENASE FROM THERMUS THERMOPHILUS HB8 : ITS THERMOSTABILITY AND STRUCTURE. | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Qu, C, Akanuma, S, Tanaka, N, Moriyama, H, Oshima, T. | | Deposit date: | 2000-10-21 | | Release date: | 2000-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, X-ray crystallography, molecular modelling and thermal stability studies of mutant enzymes at site 172 of 3-isopropylmalate dehydrogenase from Thermus thermophilus.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1FGJ
 
 | | X-RAY STRUCTURE OF HYDROXYLAMINE OXIDOREDUCTASE | | Descriptor: | HEME C, HYDROXYLAMINE OXIDOREDUCTASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tanaka, N, Igarashi, N, Moriyama, H. | | Deposit date: | 1997-03-03 | | Release date: | 1998-03-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The 2.8 A structure of hydroxylamine oxidoreductase from a nitrifying chemoautotrophic bacterium, Nitrosomonas europaea.
Nat.Struct.Biol., 4, 1997
|
|
