4G2I
 
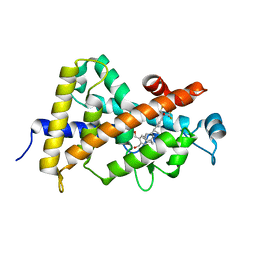 | | Structural basis for the accommodation of bis- and tris-aromatic derivatives in Vitamin D Nuclear Receptor | | Descriptor: | (3E,5E)-6-(3-{2-[3,4-bis(hydroxymethyl)phenyl]ethyl}phenyl)-1,1,1-trifluoro-2-(trifluoromethyl)octa-3,5-dien-2-ol, Vitamin D3 receptor | | Authors: | Ciesielski, F, Sato, Y, Moras, D, Rochel, N. | | Deposit date: | 2012-07-12 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the accommodation of bis- and tris-aromatic derivatives in vitamin d nuclear receptor.
J.Med.Chem., 55, 2012
|
|
4G1Y
 
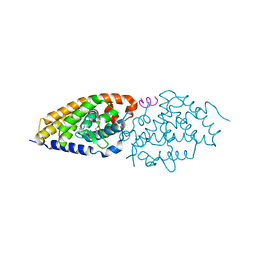 | | Structural basis for the accommodation of bis- and tris-aromatic derivatives in Vitamin D Nuclear Receptor | | Descriptor: | (4E,6Z)-7-(3-{[3,4-bis(hydroxymethyl)benzyl]oxy}phenyl)-3-ethylnona-4,6-dien-3-ol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Ciesielski, F, Sato, Y, Moras, D, Rochel, N. | | Deposit date: | 2012-07-11 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis for the accommodation of bis- and tris-aromatic derivatives in vitamin d nuclear receptor.
J.Med.Chem., 55, 2012
|
|
4G1Z
 
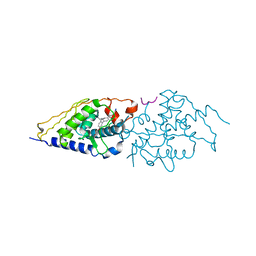 | | Structural basis for the accommodation of bis- and tris-aromatic derivatives in Vitamin D Nuclear Receptor | | Descriptor: | 3-(5'-{[3,4-bis(hydroxymethyl)benzyl]oxy}-2'-ethyl-2-propylbiphenyl-4-yl)pentan-3-ol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Ciesielski, F, Sato, Y, Moras, D, Rochel, N. | | Deposit date: | 2012-07-11 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the accommodation of bis- and tris-aromatic derivatives in vitamin d nuclear receptor.
J.Med.Chem., 55, 2012
|
|
4G2H
 
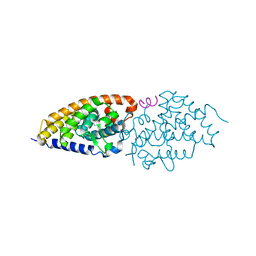 | | Structural basis for the accommodation of bis- and tris-aromatic derivatives in Vitamin D Nuclear Receptor | | Descriptor: | (3E,5E)-6-(3-{2-[3,4-bis(hydroxymethyl)phenyl]ethyl}phenyl)-1,1,1-trifluoro-2-(trifluoromethyl)octa-3,5-dien-2-ol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Ciesielski, F, Sato, Y, Moras, D, Rochel, N. | | Deposit date: | 2012-07-12 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the accommodation of bis- and tris-aromatic derivatives in vitamin d nuclear receptor.
J.Med.Chem., 55, 2012
|
|
6HN6
 
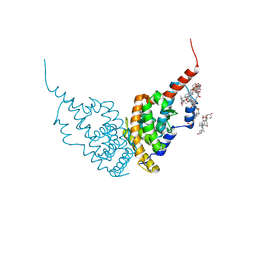 | | A revisited version of the apo structure of the ligand-binding domain of the human nuclear receptor RXR-ALPHA | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Retinoic acid receptor RXR-alpha | | Authors: | Eberhardt, J, McEwen, A.G, Bourguet, W, Moras, D, Dejaegere, A. | | Deposit date: | 2018-09-14 | | Release date: | 2019-02-20 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | A revisited version of the apo structure of the ligand-binding domain of the human nuclear receptor retinoic X receptor alpha.
Acta Crystallogr F Struct Biol Commun, 75, 2019
|
|
4LBD
 
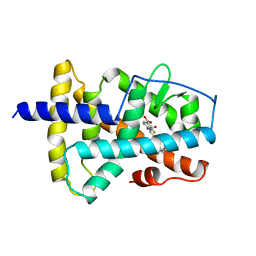 | | LIGAND-BINDING DOMAIN OF THE HUMAN RETINOIC ACID RECEPTOR GAMMA BOUND TO THE SYNTHETIC AGONIST BMS961 | | Descriptor: | 3-FLUORO-4-[2-HYDROXY-2-(5,5,8,8-TETRAMETHYL-5,6,7,8,-TETRAHYDRO-NAPHTALEN-2-YL)-ACETYLAMINO]-BENZOIC ACID, RETINOIC ACID RECEPTOR GAMMA | | Authors: | Klaholz, B.P, Renaud, J.-P, Mitschler, A, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 1998-02-04 | | Release date: | 1999-03-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational adaptation of agonists to the human nuclear receptor RAR gamma.
Nat.Struct.Biol., 5, 1998
|
|
7Q71
 
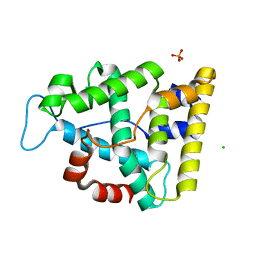 | | The crystallographic structure of the Ligand Binding domain of the NR7 nuclear receptor from the amphioxus Branchiostoma lanceolatum | | Descriptor: | CHLORIDE ION, Nuclear hormone receptor 7, PHOSPHATE ION | | Authors: | Billas, I.M.L, McEwen, A.G, Hazemann, I, Moras, D, Laudet, V. | | Deposit date: | 2021-11-09 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel nuclear receptor subfamily enlightens the origin of heterodimerization.
Bmc Biol., 20, 2022
|
|
1YZ1
 
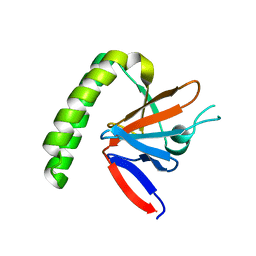 | |
1Z60
 
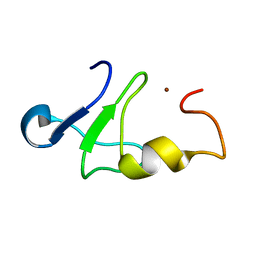 | | Solution structure of the carboxy-terminal domain of human TFIIH P44 subunit | | Descriptor: | TFIIH basal transcription factor complex p44 subunit, ZINC ION | | Authors: | Kellenberger, E, Dominguez, C, Fribourg, S, Wasielewski, E, Moras, D, Poterszman, A, Boelens, R, Kieffer, B. | | Deposit date: | 2005-03-21 | | Release date: | 2005-04-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal domain of TFIIH P44 subunit reveals a novel type of C4C4 ring domain involved in protein-protein interactions.
J.Biol.Chem., 280, 2005
|
|
1VTQ
 
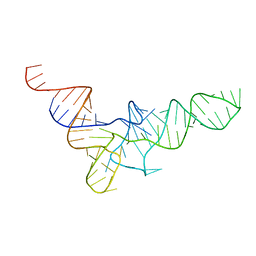 | | THREE-DIMENSIONAL STRUCTURE OF YEAST T-RNA-ASP. I. STRUCTURE DETERMINATION | | Descriptor: | T-RNA-ASP | | Authors: | Comarmond, M.B, Giege, R, Thierry, J.C, Moras, D, Fischer, J. | | Deposit date: | 1985-06-11 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three-Dimensional Structure of Yeast T-RNA-ASP. I. Structure Determination
Acta Crystallogr.,Sect.B, 42, 1986
|
|
1FCY
 
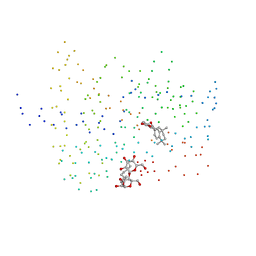 | | ISOTYPE SELECTIVITY OF THE HUMAN RETINOIC ACID NUCLEAR RECEPTOR HRAR: THE COMPLEX WITH THE RARBETA/GAMMA-SELECTIVE RETINOID CD564 | | Descriptor: | 6-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRO-NAPHTALENE-2-CARBONYL)-NAPHTALENE-2-CARBOXYLIC ACID, DODECYL-ALPHA-D-MALTOSIDE, RETINOIC ACID RECEPTOR GAMMA-1 | | Authors: | Klaholz, B.P, Mitschler, A, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-07-19 | | Release date: | 2000-09-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for isotype selectivity of the human retinoic acid nuclear receptor.
J.Mol.Biol., 302, 2000
|
|
1FD0
 
 | | ISOTYPE SELECTIVITY OF THE HUMAN RETINOIC ACID NUCLEAR RECEPTOR HRAR: THE COMPLEX WITH THE RARGAMMA-SELECTIVE RETINOID SR11254 | | Descriptor: | 6-[HYDROXYIMINO-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRO-NAPHTALEN-2-YL)-METHYL]-NAPHTALENE-2-CARBOXYLIC ACID, DODECYL-ALPHA-D-MALTOSIDE, RETINOIC ACID RECEPTOR GAMMA-1 | | Authors: | Klaholz, B.P, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-07-19 | | Release date: | 2002-09-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | C-H...O hydrogen bonds in the nuclear receptor RARgamma--a potential tool for drug selectivity.
Structure, 10, 2002
|
|
1FCZ
 
 | | ISOTYPE SELECTIVITY OF THE HUMAN RETINOIC ACID NUCLEAR RECEPTOR HRAR: THE COMPLEX WITH THE PANAGONIST RETINOID BMS181156 | | Descriptor: | 4-[3-OXO-3-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRO-NAPHTHALEN-2-YL)-PROPENYL]-BENZOIC ACID, DODECYL-ALPHA-D-MALTOSIDE, RETINOIC ACID RECEPTOR GAMMA-1 | | Authors: | Klaholz, B.P, Mitschler, A, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-07-19 | | Release date: | 2000-09-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural basis for isotype selectivity of the human retinoic acid nuclear receptor.
J.Mol.Biol., 302, 2000
|
|
1FCX
 
 | | ISOTYPE SELECTIVITY OF THE HUMAN RETINOIC ACID NUCLEAR RECEPTOR HRAR: THE COMPLEX WITH THE RARGAMMA-SELECTIVE RETINOID BMS184394 | | Descriptor: | 6-[HYDROXY-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRO-NAPHTALEN-2-YL)-METHYL]-NAPHTALENE-2-CARBOXYLIC ACID, DODECYL-ALPHA-D-MALTOSIDE, RETINOIC ACID RECEPTOR GAMMA-1 | | Authors: | Klaholz, B.P, Mitschler, A, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-07-19 | | Release date: | 2000-09-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural basis for isotype selectivity of the human retinoic acid nuclear receptor.
J.Mol.Biol., 302, 2000
|
|
1GGM
 
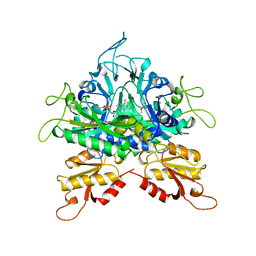 | |
3LBD
 
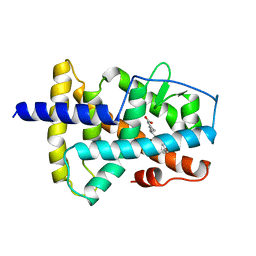 | | LIGAND-BINDING DOMAIN OF THE HUMAN RETINOIC ACID RECEPTOR GAMMA BOUND TO 9-CIS RETINOIC ACID | | Descriptor: | (9cis)-retinoic acid, RETINOIC ACID RECEPTOR GAMMA | | Authors: | Klaholz, B.P, Renaud, J.-P, Mitschler, A, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 1998-02-04 | | Release date: | 1999-03-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational adaptation of agonists to the human nuclear receptor RAR gamma.
Nat.Struct.Biol., 5, 1998
|
|
3DR1
 
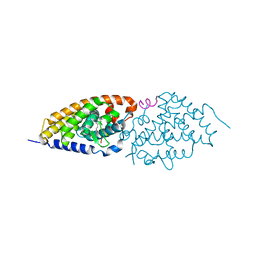 | | Side-chain fluorine atoms of non-steroidal vitamin D3 analogs stabilize helix 12 of vitamin D receptor | | Descriptor: | (1R,3R)-5-[(2E)-3-{(1S,3R)-2,2,3-trimethyl-3-[6,6,6-trifluoro-5-hydroxy-5-(trifluoromethyl)hex-3-yn-1-yl]cyclopentyl}prop-2-en-1-ylidene]cyclohexane-1,3-diol, MAGNESIUM ION, SRC-1 (LXXLL motif) from Nuclear receptor coactivator 1, ... | | Authors: | Sato, Y, Rochel, N, Moras, D. | | Deposit date: | 2008-07-10 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Superagonistic fluorinated vitamin D3 analogs stabilize helix 12 of the vitamin D receptor.
Chem.Biol., 15, 2008
|
|
3EYB
 
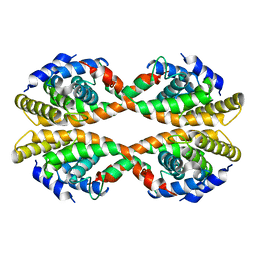 | |
3TKC
 
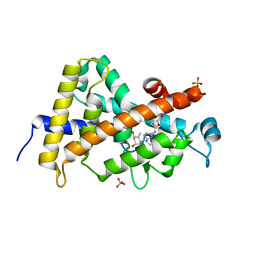 | | Design, Synthesis, Evaluation and Structure of Vitamin D Analogues with Furan Side Chains | | Descriptor: | (1S,3R,5Z,7E,14beta,17alpha,20S)-20-[5-(1-hydroxy-1-methylethyl)furan-2-yl]-9,10-secopregna-5,7,10-triene-1,3-diol, SULFATE ION, Vitamin D3 receptor | | Authors: | Huet, T, Moras, D, Rochel, N. | | Deposit date: | 2011-08-26 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Design, synthesis, evaluation, and structure of vitamin D analogues with furan side chains.
Chemistry, 18, 2012
|
|
4TRA
 
 | |
4AWL
 
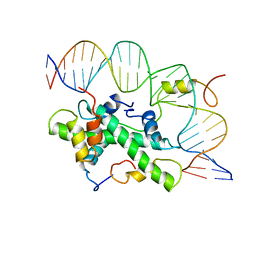 | | The NF-Y transcription factor is structurally and functionally a sequence specific histone | | Descriptor: | HSP70 PROMOTER FRAGMENT, NUCLEAR TRANSCRIPTION FACTOR Y SUBUNIT ALPHA, NUCLEAR TRANSCRIPTION FACTOR Y SUBUNIT BETA, ... | | Authors: | Nardini, M, Gnesutta, N, Donati, G, Gatta, R, Forni, C, Fossati, A, Vonrhein, C, Moras, D, Romier, C, Mantovani, R, Bolognesi, M. | | Deposit date: | 2012-06-04 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Sequence-Specific Transcription Factor NF-Y Displays Histone-Like DNA Binding and H2B-Like Ubiquitination.
Cell(Cambridge,Mass.), 152, 2013
|
|
1EXX
 
 | | ENANTIOMER DISCRIMINATION ILLUSTRATED BY CRYSTAL STRUCTURES OF THE HUMAN RETINOIC ACID RECEPTOR HRARGAMMA LIGAND BINDING DOMAIN: THE COMPLEX WITH THE INACTIVE S-ENANTIOMER BMS270395. | | Descriptor: | 3-FLUORO-4-[2-HYDROXY-2-(5,5,8,8-TETRAMETHYL-5,6,7,8,-TETRAHYDRO-NAPHTALEN-2-YL)-ACETYLAMINO]-BENZOIC ACID, DODECYL-ALPHA-D-MALTOSIDE, RETINOIC ACID RECEPTOR GAMMA-2 | | Authors: | Klaholz, B.P, Mitschler, A, Belema, M, Zusi, C, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-05-05 | | Release date: | 2000-06-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Enantiomer discrimination illustrated by high-resolution crystal structures of the human nuclear receptor hRARgamma.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1FYF
 
 | | CRYSTAL STRUCTURE OF A TRUNCATED FORM OF THREONYL-TRNA SYNTHETASE COMPLEXED WITH A SERYL ADENYLATE ANALOG | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, THREONYL-TRNA SYNTHETASE, ZINC ION | | Authors: | Sankaranarayanan, R, Dock-Bregeon, A.C, Moras, D. | | Deposit date: | 2000-09-29 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Transfer RNA-mediated editing in threonyl-tRNA synthetase. The class II solution to the double discrimination problem.
Cell(Cambridge,Mass.), 103, 2000
|
|
2QEN
 
 | | The walker-type atpase paby2304 of pyrococcus abyssi | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, WALKER-TYPE ATPASE | | Authors: | Uhring, M, Bey, G, Lecompte, O, Moras, D, Poch, O, Cavarelli, J. | | Deposit date: | 2007-06-26 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The walker-type atpase paby2304 of pyrococcus abyssi
To be Published
|
|
1YJE
 
 | | Crystal structure of the rNGFI-B ligand-binding domain | | Descriptor: | Orphan nuclear receptor NR4A1 | | Authors: | Flaig, R, Greschik, H, Peluso-Iltis, C, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-01-14 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the cell-specific activities of the NGFI-B and the Nurr1 ligand-binding domain.
J.Biol.Chem., 280, 2005
|
|
